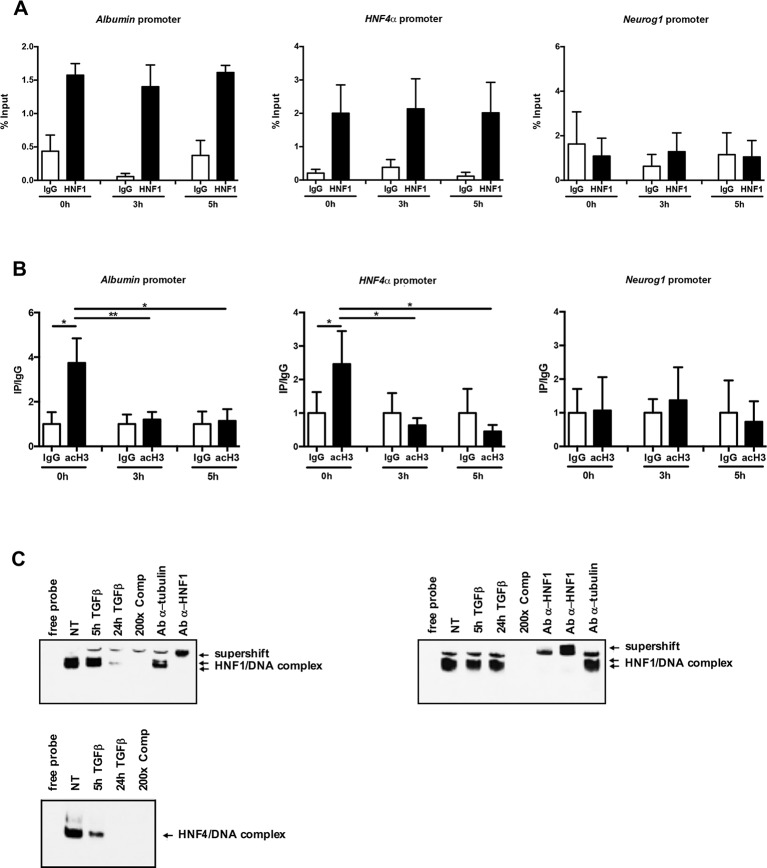
Figure 3.
(A) HNF1α DNA binding activity after TGFβ treatment. qPCR analysis of chromatin immunoprecipitated from hepatocytes with anti-HNF1α antibody was performed. HNF1α consensus regions embedded in the indicated HNF1α target gene promoters were analyzed. A HNF1α nonbound region of Neurogenin 1 promoter was utilized as negative control. Data are normalized to total chromatin input and background (control immunoprecipitation with IgG) and expressed as % input. Mean ± SEM of qPCR data obtained in triplicate from three independent experiments are reported. (B) Analysis of acetyl histone H3 by chromatin immunoprecipitation assay in untreated and TGFβ-treated hepatocytes. qPCR analysis of chromatin immunoprecipitated from hepatocytes with anti-acetyl H3 antibody was performed. HNF1α consensus regions embedded in the indicated HNF1α target gene promoters were analyzed. A HNF1α nonbound region of Neurogenin 1 promoter was utilized as negative control. Data are normalized to total chromatin input and background (control immunoprecipitation with IgG) and expressed as (Ip/IgG) % input. The mean ± SEM of qPCR data obtained in triplicate from five independent experiments are reported. *p <0.05, **p <0.01. (C) Electrophoretic mobility shift assay (EMSA) assays. Nuclear extracts from untreated (NT) or TGFβ-treated (for 5 or 24 h) parental (left, upper panel) or HNF1α-overexpressing hepatocytes (right, upper panel) were analyzed for the binding to HNF1α consensus site within the murine HNF4α promoter. The presence of the HNF1α protein in the protein/DNA complexes was revealed by the band supershift obtained with the addition of two different anti-HNF1α antibodies. A 200-fold excess of unlabeled oligonucleotide and the antitubulin antibody were added to the untreated extracts to test the binding specificity. As control, HNF4α DNA binding activity to its consensus site within the ApoC3 promoter was analyzed by EMSA in the same extracts (lower panel).