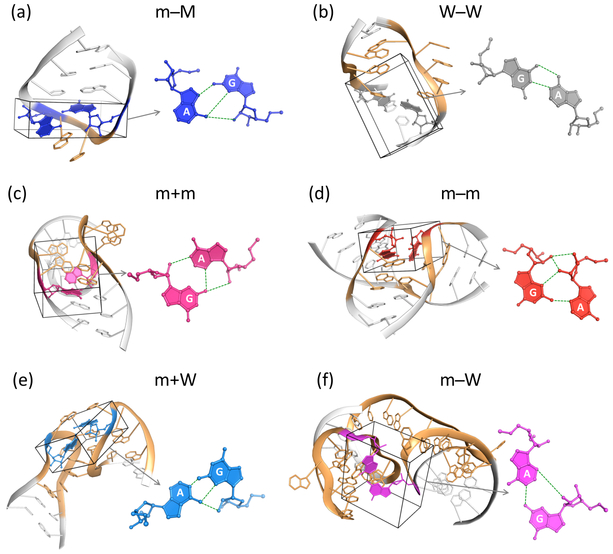
Figure 2.
Molecular images of RNA secondary structural motifs incorporating each of the dominant modes of G·A base pairing. Motifs correspond to one of the common settings of the designated pairs: (a) sheared m−M pair closing the GNRA tetraloop at the end of the P4 helix of the glmS ribozyme bound to glucosamine 6-phosphate;35 (b) imino W−W pair at the end of an asymmetric internal loop in the Thermus thermophilus 70S ribosome in complex with the hibernation factor pY;36 (c) m+m pair linking the G in a double-helical stem and the A in an internal loop of the complex of Thermus thermophilus ribosomal protein L1 with a fragment of the L1 RNA from Methanoccocus vannielii;37 (d) m−m pair joining a stem and single-stranded fragment within a three-way junction of the Thermoanaerobacter tengcongenesis ydaO riboswitch bound to cyclic di-AMP;38 (e) m+W pair connecting the D and T hairpin loops of yeast initiator transfer RNA;39 (f) m−W pair linking two hairpin loops of the 5S rRNA within the structure of the hibernating 100S ribosome dimer from pathogenic Staphylococcus aureus.40 G·A pairs color-coded as in Figure 1 and shown at the nucleotide level within each structural element and in a separate local depiction to the right of each example. Loops depicted in gold and canonical base pairs at the ends of loops or within double-helical stems in white. Ribbons connect phosphorus atoms in successive nucleotides. See Table S3 for respective Protein Data Bank identifiers, chain names, and residue numbers of the depicted pairs and Figure S2 for simple secondary structural diagrams of the associated motifs.