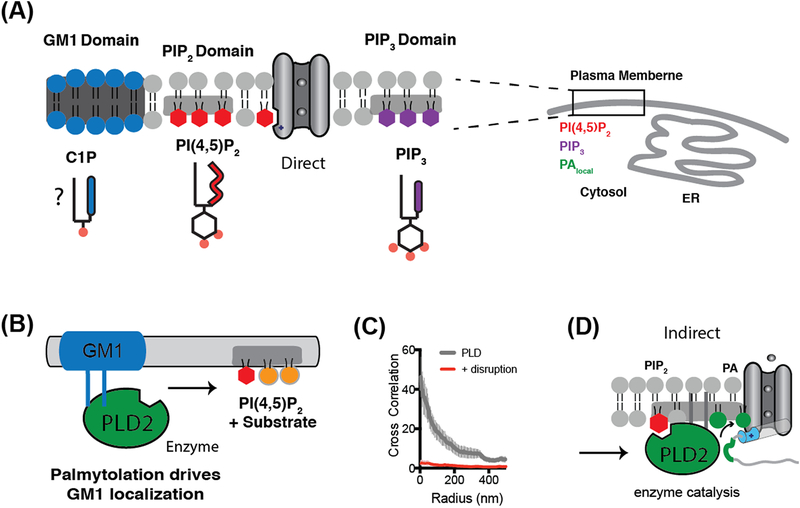
Figure 2. Lipid partitioning and clustering gives rise to lipid heterogeneity.
(A) Lipids partition into at least three domains or regions of the cell membrane. The regulation of a channel depends on its partitioning with agonizing or antagonizing anionic signaling lipids (e.g. PIP2 and PIP3). Ceramide-1-phosphated (C1P) is an anionic ceramide comprised of a phosphate head group (red dot) similar to phosphatidic acid and expected to localize to GM1 domains. The endoplasmic reticulum (ER) is shown for reference and scale. (B) PLD2 is activated by substrate presentation. GM1 domains sequester PLD2 away from its substrate phosphatidylcholine (PC, orange circles). Increases in PIP2 attract PLD2 away from the GM1 domains localizing the enzyme near PC allowing for hydrolysis. (C) Cross correlation analysis of PLD association with GM1 domains before and after disruption with methyl-beta-cyclodextrin. After disruption, PLD fails to correlate with GM1 at any distance (radius in nm, error bars shown as standard error of the mean (n=7–8). (D) PIP2 activation of PLD2 can then indirectly activate TREK-1. The enzyme localizes to the disordered C-terminus (grey strand) of TREK-1 through a PLD2 binding site (green segment). PLD2 produces anionic lipid phosphatidic acid (PA) (green lipid) which binds to lipid binding site (blue cylinder) which opens the channel. This indirect mechanism contrasts with PIP2 directly activating a channel shown in (A).