Figure 6.
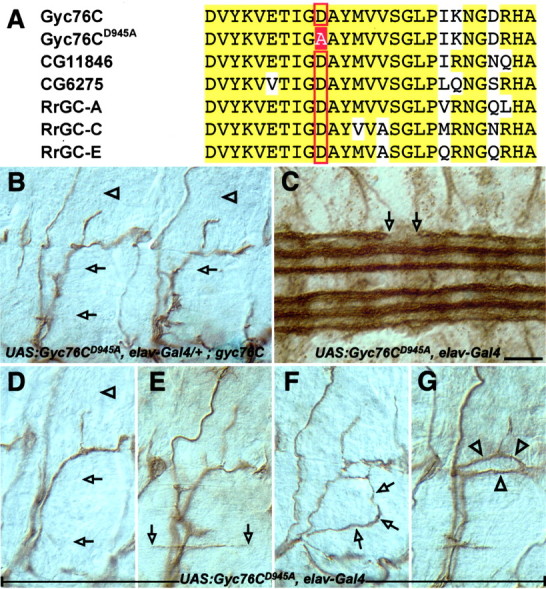
A catalytically inactive form of Gyc76C fails to rescue the gyc76C mutant phenotype and perturbs motor and CNS axon guidance. A, Alignment of a portion of the catalytic regions of three Drosophila and three rat rGCs. The red box indicates the catalytic aspartate (D945) that was converted to alanine to make the Gyc76CD945A transgene. B-G, Filleted preparations of late stage 16 embryos stained with the anti-fasciclin II monoclonal antibody to label motor and CNS axons, as in Figure 4. B, An embryo expressing the mutant Gyc76CD945A in all neurons in a mutant gyc76C background exhibits the same defects observed in gyc76C homozygous mutants. C-G, Overexpressing the mutant Gyc76CD945A in all neurons in a wild-type background disrupts normal CNS and motor axon pathfinding. C, An embryo overexpressing the mutant Gyc76CD945A in a wild-type background shows an interruption of a CNS axon tract with a large gap in the outermost bundle of longitudinal axons within the CNS (open arrows). D, A wild-type embryo overexpressing Gyc76CD945A in all neurons displays LOF-like phenotypes in which axons of the ISNb (open arrowheads) and SNa (open arrows) fail to reach their proper targets. E-G, Overexpressing Gyc76CD945A in all neurons in a wild-type background also produces novel phenotypes. E, The RP3 axon (open arrowheads) extends exuberantly in the cleft between muscles 6 and 7. The ISNb (F, open arrows) and SNa (G, open arrowheads) axon bundles wander inappropriately as they extend toward their targets. Scale bar: B-G, 10 μm.
