Figure 1.
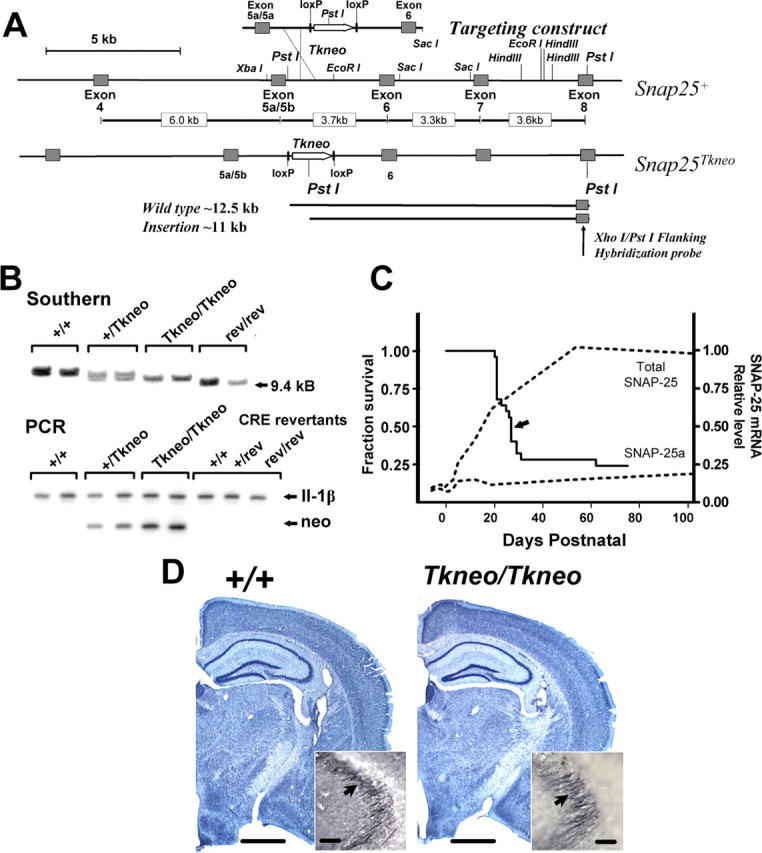
Targeted insertion of Tkneo selection transgene downstream of exon 5a and 5b in the Snap25 gene results in postnatal lethality. A, Schematic diagram demonstrating the generation and structure of the mutation Snap25Tkneo. The floxed Tkneo selection gene was inserted at the EcoRI site between the tandemly arranged exons 5a and 5b and exon 6 in a targeting construct generated from the wild-type gene (Snap25+) described in Washbourne et al. (2002). In the targeting construct, a PvuI-HindIII fragment containing exon 5a and 5b sequences was exchanged for an exon 5a-5a sequence produced by multiplex PCR (C. Bark, unpublished data). Sequence analysis of genomic PCR products obtained from DNA of homozygous mutant mice demonstrated that exon 5a and 5b in fact were intact in Snap25Tkneo mutant mice (see Material and Methods), indicating that the 5′ site of homologous recombination was downstream of exon 5. The bottom of the panel depicts the 3′ flanking XhoI-PstI fragment used to screen ES cell and genomic DNAs for homologous recombination by Southern blotting and the expected PstI fragments detected from wild-type (∼12.5 kb) and the targeted mutant (∼11 kb) alleles. A more representative diagram of the genomic structure of the Snap25 gene locus with the lengths of the respective intron sequences is shown between the maps of the endogenous Snap25+ and mutant Snap25Tkneo alleles. B, Representative genotyping by Southern blot of PstI restriction fragment length and by a genomic PCR assay that compares products of semiquantitative amplification of the inserted TKneo gene (neo) and of IL-1β, a flanking control gene on mouse chromosome 2. C, Survival curve demonstrating the postnatal lethality of homozygous Snap25Tkneo mutants (n = 25). Seven of 25 mice died before weaning, which is represented by the sharp drop at P21, and 6 of 25 survived >75 d; the median age of survival of P27 is indicated by an arrow. No remarkable postnatal lethality was observed for heterozygote or revertant mice, and their survival appears comparable with wild type. For comparison, the expression profiles of SNAP-25a and total SNAP-25 mRNAs, modified from Bark et al. (1995), are shown (dotted lines). D, Nissl staining reveals grossly normal morphology of mutant adult (PN78) brains (right, Tkneo/Tkneo) compared with wild-type (left, +/+) littermates; note the well structured development of the hippocampal formation in the mutant. Inset, Immunohistochemical staining for SNAP-25 of mossy fiber presynaptic terminals adjacent to the CA3 region of hippocampus (arrows) shows the characteristic accumulation of SNAP-25 in mutant mice, comparable with wild type. Scale bars: D, 1 mm; inset in D, 50 μm.
