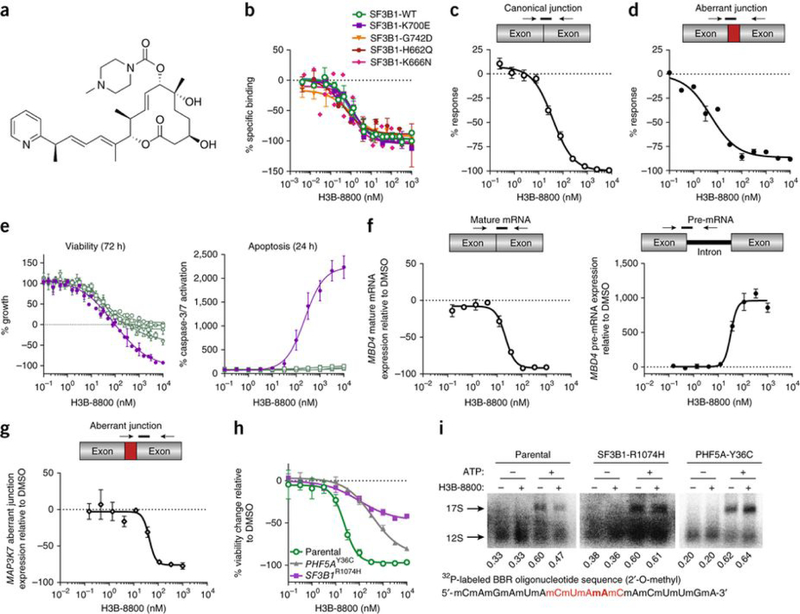
Figure 1.
H3B-8800 modulates splicing of WT and mutant SF3B1 spliceosomes in vitro and preferentially kills SF3B1-mutant cells. (a) The chemical structure of H3B-8800. (b) Competitive binding of increasing concentrations of H3B-8800 to SF3b complexes isolated from cells overexpressing WT or mutant SF3B1 in the presence of 1 nM pladienolide B. The y axis represents the percentage change (% response) of specific binding relative to the DMSO control of 0%. Data are represented as mean ± s.d.; n = 3 experimental replicates. (c,d) Quantification of canonical (c) and aberrant (d) junctions using the Ad2-ZDHHC16 pre-mRNA substrate, in which the original Ad2 intron was substituted with intron 9 of ZDHHC16, in an in vitro splicing assay using nuclear extracts from 293FT cells overexpressing SF3B1K700E and treated with increasing concentrations of H3B-8800. The y axis represents the percent response relative to the DMSO control (0%). Data are represented as mean ± s.d.; n = 3 technical replicates. (e) Viability and apoptosis of pancreatic cells bearing WT (Panc10.05, HAPFII, CFPAC1, and Panc04.03 in green) or mutant (Panc05.04 in violet) SF3B1 treated with increasing concentrations of H3B-8800. Data are represented as mean ± s.d.; n = 3 technical replicates. (f,g) Effect of increasing concentrations of H3B-8800 on splicing of MBD4 mature mRNA (open circles) and MBD4 pre-mRNA (black circles) (f) and MAP3K7 aberrant junction expression (open diamonds) (g) in Panc05.04 cells as quantified by TLDA after 6 h of treatment with H3B-8800. Data are represented as mean ± s.e.m.; n = 3 technical replicates. (h) Viability of HCT116 parental cells and clones carrying the pladienolide-resistance mutation SF3B1R1074H or PHF5AY36C. Data are represented as mean± s.d.; n = 3 technical replicates. (i) Binding of U2 snRNA to the canonical branchpoint sequence (2′-O-methyl BBR oligonucleotide in red with the branchpoint adenosine bolded) in nuclear extracts from the same HCT116 cells as in h. Nuclear extracts were incubated in the presence or absence of ATP and H3B-8800; band quantification is shown below each lane. The functional 17S U2 snRNP was formed via ATP-dependent binding of the SF3b complex to the nonfunctional 12S U2 snRNP particle.