Abstract
Polymorphonuclear cells (PMNs or neutrophils) are the most abundant leukocyte in humans and represent an essential component of the innate immune system. The ability of neutrophils to initiate an immediate and non-specific host response against invading microbial species is key to determining the outcome of infection. Neutrophils produce and secrete a plethora of immunomodulatory proteins, including major granule proteins and cytokines, as well as various enzymes, which regulate adherence, phagocytosis, chemotaxis, and cell survival. Historically, characterization of neutrophils and their roles during infection have relied on genetic and phenotypic analyses, as well as biochemical assays. However, recent advances in mass spectrometry-based proteomic workflows and technological platforms have supported the comprehensive profiling of neutrophil-associated immune responses in consideration of cellular factors and secreted proteins. Given the critical role of neutrophils in maintaining and regulating innate immune function, comprehensive profiling of their response to infection is imperative to ensuring host survival. Here, we briefly discuss the role of neutrophils in host defense and describe methods to purify neutrophils from murine samples and comprehensively profile their proteomes.
Basic Protocol 1: Purification of mouse bone marrow-derived neutrophils
Basic Protocol 2: Flow cytometry analysis of purified neutrophils
Basic Protocol 3: Protein extraction from neutrophils
Basic Protocol 4: Proteomic profiling of neutrophils
Keywords: Mass spectrometry, quantitative proteomics, immune cells, polymorphonuclear neutrophils, flow cytometry
INTRODUCTION:
Recent technological advances in the field of mass spectrometry-based proteomics have improved instrument sensitivity, accuracy, and robustness for analyzing complex proteomes1,2. Additionally, improvements to the accessibility and user-friendly designs of bioinformatic pipelines promote consistent and reliable analysis of these complex data sets3,4. Discovery-based proteomics, employing a bottom-up strategy, which relies on the enzymatic digestion of proteins into peptides, offers a uniquely unbiased approach for profiling of comprehensive cellular proteomes and defining communication patterns among cells, including microbial and immune cell systems. For leukocytes, and specifically, polymorphonuclear neutrophils (PMNs) the presence of many cellular enzymes presents challenges associated with rapid protein degradation and may limit proteome coverage when analyzing samples5. To overcome this challenge, we have devised a step-by-step protocol for the comprehensive and quantitative profiling of neutrophil proteomes. Moreover, analysis of the neutrophil proteomes provides information on differences between PMNs derived from unique mouse strains and highlights specific proteins and pathways involved in neutrophil response to infection6,7.
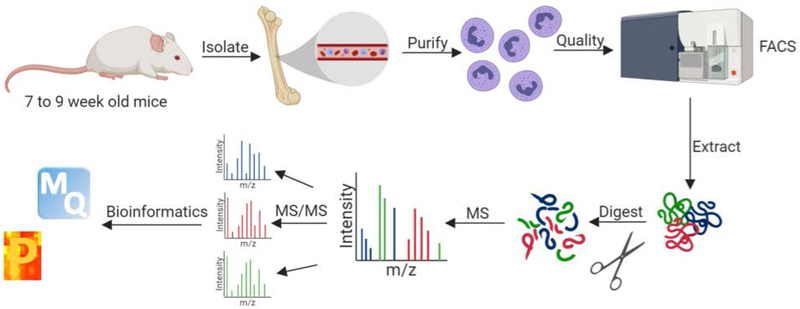
The first two protocols (Basic Protocol 1 & 2) describe the isolation and purification of bone marrow-derived neutrophils from mice and assessment of purity using flow cytometry. The neutrophil samples are then applied to the Basic Protocol 3 for protein extraction using an in-solution digestion approach with Lys-C and trypsin proteases to cut proteins into peptides for identification using liquid chromatography coupled to mass spectrometry (LC-MS/MS), as described in Basic Protocol 4. An overview of the pipeline described herein in presented in Figure 1. Beyond neutrophils, this protocol extends to the comprehensive profiling of other innate immune cells (e.g. macrophages). Overall, our approach enables us to define fundamental differences of neutrophils at baseline and explore mechanisms involved in the effective clearance of foreign invaders (e.g. bacterial cells) during infection.
Figure 1: Overview of workflow for MS-based proteomics profiling of neutrophils.
Neutrophils are isolated and purified from bone marrow of 7 to 9-week-old male or female mice. The quality of the purification is assessed by FACS. Proteins are extracted by chemical (e.g., urea) and mechanical (e.g., sonication) cell disruption and enzymatically digested with Lys-C and trypsin proteases. The purified peptides are measured in the first MS scan and peptide fragmentation patterns are observed in the second MS scan (MS/MS). The data is processed and analyzed using the publicly-available MaxQuant and Perseus platforms. Figure generated with Biorender.com.
BASIC PROTOCOL 1
Purification of mouse bone marrow-derived neutrophils
The following protocols describes isolation of neutrophils from murine bone marrow via negative selection approaches. This is our primary approach to generate purified neutrophils for functional studies. Our prior experience with murine neutrophil isolations via density gradients and positive selection, yielded more artificially activated neutrophils. Consequently, those approaches resulted in significantly lower neutrophil yield per mouse.
Materials:
Donor mice: C57BL6/N or Swiss Webster (SW), 7 to 9 weeks old, male or female (Taconic Farms)
Neutrophil isolation media (See Reagents and Solutions)
10X Phosphate buffered saline (PBS) (Boston BioProducts, cat no BM220)
Bovine serum albumin (BSA) V (American Bioanalytical, cat no. 00448–00100)
2 mM EDTA (Ambio, cat no. AM9260G)
Biotin-antibody cocktail, component of the MojoSort neutrophil kit (Biolegend, cat no. 480058)
0.4% Tryptan Blue solution (Sigma, cat no. T8154)
27G 1/4 needles (BD Needle, cat no. 305136)
20 ml syringes (BD, cat no. BD302830G)
70 μm filter (Fisherbrand, cat no. 22363548)
15 ml polypropylene round bottom tubes (e.g. BD Falcon)
Petri-dishes with clear lid (Fisherbrand, cat no. FB0875714G)
50 ml conical centrifuge tubes (e.g. BD Falcon)
5 ml polypropylene round bottom tube (e.g. BD Falcon)
MojoSort Magnet (Biolegend, cat no. 480019/480020) or compatible
Refrigerated centrifuge (Eppendorf, model 5804R)
CO2 euthanasia chamber
Transmission light microscope (Olympus CX23)
Sacrifice the mice by placing them in CO2 chamber and utilizing a secondary method for euthanasia such as cervical dislocation
Dissect hind legs, place them on ice, remove all muscle tissue, place the cleaned bones in Neutrophil isolation media, in round petri-dishes.
-
Flush the bone marrow using 27G needle into 15 ml tube.
Typically, use 1ml of Neutrophil isolation media per bone. When flushed thoroughly, the bone tissue appears white.
Filter cell suspensions through a 70 μm filter into 50 ml falcon tubes.
Centrifuge at 410 × g (1500 rpm) for 10 min at 4°C.
Decant the supernatants.
Resuspend cells in Neutrophil isolation media.
Spin 117 × g (800 rpm) for 8 min at 4°C.
Resuspend in 1 ml of Neutrophil isolation media.
-
Count cells using tryptan blue exclusion approach.
Typical yields are 40–50×106 cells/mouse
Place 100 μl of cell suspension (containing 107 bone marrow cells), into 5 ml polypropylene round bottom tube, add 10 μl of biotin-antibody cocktail. Scale accordingly.
Mix and incubate for 15 min on ice.
Wash the cells by topping up the sample tube with 4 ml of Neutrophil isolation media.
Centrifuge at 300 × g (1200 rpm) for 5 min at 4°C.
Discard the supernatant and resuspend cells to the original volume (e.g., 100 μl) with Neutrophil isolation media.
Vortex the magnetic particles for 15 sec.
-
Add 10 μl of magnetic particles to sample.
The volume of particles depends on the initial number of bone marrow cells and antibody used in step 13. Scale accordingly, as per manufacturer’s instructions.
Mix and incubate for 15 min on ice (2–8°C).
Wash the cells by topping up the sample tube with 4ml of Neutrophil isolation media and centrifuge 300 × g for 5 min at 4°C.
Discard the supernatant.
-
Add 2 ml of Neutrophil isolation media
If you observe aggregates, disrupt by pipetting.
Place in the magnet for 5 min.
Pour out the unlabeled fraction and keep the supernatant. Store on ice.
Resuspend the labeled cells in 2 mL of Neutrophil isolation media.
Place the tube in the magnet for additional 5 min.
Pour out the unlabeled fraction and pool with the previously collected unlabeled cells.
-
Place the pooled unlabeled fraction in the magnet for additional 5 min.
This step ensures appropriate enrichment of neutrophils.
Pour out the unlabeled fraction and keep the purified neutrophils.
-
As done in step 10.
The typical yield per mouse upon completion of the protocol is 5–7×106 purified neutrophils. Cells should not stored for extensive periods of time; it is advised to proceed speedily with functional analysis such as Basic Protocol 2 or Basic Protocol 3.
Basic Protocol 2:
Flow cytometry analysis of purified neutrophils
We typically use a flow cytometry analysis to determine PMN purity and viability. A combination of forward scatter (FSC), side scatter (SSC), Ly6G, and CD11b staining provides a robust and quick approach to estimate total PMN yields.
Materials:
Purified neutrophils and isolated bone marrow cells (Basic protocol 1, step 29)
Flow cytometry buffer (PBS, 5% FBS)
10X PBS (Boston BioProducts, cat no. BM220)
Fetal Bovine Serum (ThermoFisher, cat no. 26140)
FC block: rat anti-mouse CD16/CD32 clone 2.4G2 in cell staining buffer (Biolegend, cat no. 101302)
Fluorescently labeled antibodies (see Table 1)
Table 1:
Antibodies for flow cytometry analysis of bone marrow-derived neutrophils
| CD11b-FITC | Biolegend, cat no 101205 | 0.25 μg/1×106 cells |
| Ly6G-PE | Biolegend, cat no 127607 | 0.25 (μg/1×106 cells |
Propidium iodide (BD Pharmingen, cat no. 55211)
5 ml polypropylene round bottom tubes (e.g. BD Falcon)
Refrigerated centrifuge (Eppendorf, model 5804R)
Fluorescence-activated cell sorting (FACS) Calibur or equivalent
CellQuest analysis software or equivalent (e.g. FlowJo)
Resuspend 0.5–1×106 neutrophils (Basic Protocol 1, step 29) in 100 μl of flow cytometry buffer and add appropriate amount of Fc block (typically diluted 1/100) as per manufacturer’s instruction.
Incubate for 30 min at 4°C.
-
Add antibody combinations (e.g., CD11b-FITC, Ly6G-PE) and incubate samples for 30 min in the dark at 4°C.
Make sure you set up individually stained control samples and non-stained control samples.
Wash twice with at least 2 ml of flow cytometry buffer by centrifuging at 300 × g for 5 min.
-
Resuspend in 500 μl of flow cytometry buffer and analyze on FACS Calibur.
Live cells should be analyzed immediately. We typically do not fix cells for subsequent analysis.
Plot side scatter (SSC) and forward scatter (FSC) and ensure that all fluorochromes are compensated. Gate on all cells. Acquire 1×105 events.
-
Compare the purified neutrophil samples with the non-purified bone marrow suspension by gating on the CD11b+ cells and determining the percent of CD11b+Ly6G+ out of the total viable cells (PI-).
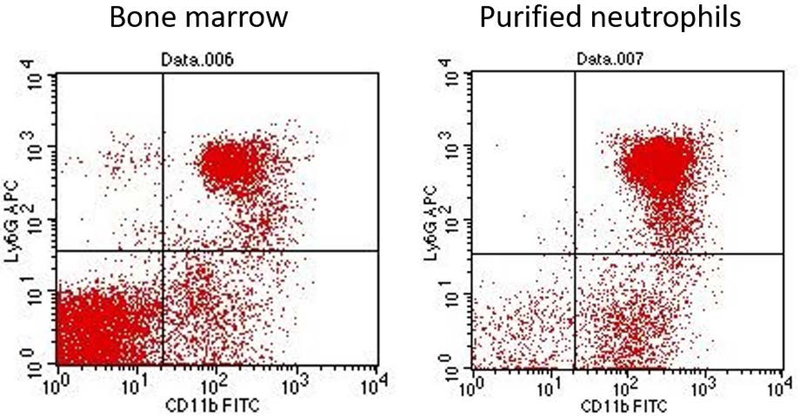
After purification, PMNs are 90% viable and present about 80% of the total cell population (Figure 2).
Figure 2: Representative flow diagram plots of viable bone marrow cells stained for neutrophil markers CD11b and Ly6G and purified PMNs.
Bone marrow cells and purified neutrophils (StemCell purification kit) were stained and analyzed in Basic Protocol 2. The figure shows significant enrichment for neutrophils in the purified population, where the Ly6G+CD11b+ cells represent 80% of the total cells.
BASIC PROTOCOL 3
Protein extraction from neutrophils
Protein extraction methods vary greatly in their cell lysis (e.g. mechanical, chemical), protein collection (e.g. solubilization, precipitation), and enzymatic digestion (e.g. protease) steps. We have explored the use of probe sonication combined with acetone precipitation for extraction of proteins from neutrophils and find the technique to be excessive for the cell type. Therefore, we have revised our extraction protocol and define an in-solution digestion method below, which relies less on mechanical disruption of cells and reduces protein loss associated with unnecessary sample handling. Although this protocol focuses on protein extraction from neutrophils, our approach can be applied to other mammalian cells (e.g. macrophages) and culture supernatants.
Materials:
PMNs (see Basic Protocols 1 and 2)
1X PBS, pH 7.4 (ThermoFisher, cat no. 10010023)
8 M Urea (Sigma-Aldrich, cat no. U1250)
40 mM HEPES (Sigma-Aldrich, cat no. H4034)
1 M Dithiothreitol (DTT) (Sigma-Aldrich, cat no. D0632)
0.55 M Iodoacetamide (IAA) (Sigma-Aldrich, cat no. I6125)
Ice
Lys-C protease (Pierce, cat no. 90307)
Trypsin protease (Pierce, cat no. 90057)
Stopping solution: 20% MS-grade acetonitrile (Pierce, cat no. TS-51101) and 6% trifluoroacetic acid (Sigma-Aldrich, cat no. 302031)
15 ml conical polypropylene centrifuge tubes (e.g. Corning, cat no. C352196)
1.5 ml microcentrifuge tubes (e.g. Eppendorf LoBind microcentrifuge tubes, cat no. 022431081)
Ultrasonic bath (e.g. Bransonic CPXH 2.5 L, digital timer, stored in a cold room) or comparable
Centrifuge (Eppendorf, model 5430)
-
Resuspend 2×106 PMNs in 200 μl of PBS in 1.5 ml microcentrifuge tube.
More PMNs can be used, but this is the recommended minimum. Adjust volume of PBS, as needed, to immerse cell pellet.
-
Add 1/3 volume of 8 M Urea & 40 mM HEPES.
The 8M Urea + 40 mM HEPES is prepared fresh in 15 ml conical polypropylene centrifuge tube and discarded after single use. Final Urea concentration is approximately 2.5 M to prevent inhibition of protease activity in subsequent steps.
Sonicate mixture for 15 cycles of 30s on/30s off in cold, circulating ultrasonic bath housed in a 4°C room.
-
Add 1:100 (v:v) of 1 M DTT (final concentration of 10 mM DTT) and mix with pipetting.
Stock solution of 1 M DTT can be prepared in bulk, flash frozen, and stored at −20°C until needed; discard after single use.
Incubate for 30 min at room temperature.
-
Add 1:10 (v:v) of 0.55 M IAA (final concentration of 55 mM IAA) and mix with pipetting.
To minimize degradation of IAA, perform step 5 in the dark. Stock solution of 0.55 M IAA can be prepared in bulk, flash frozen, and stored in the dark at −20°C until needed; discard after single use.
Incubate for 20 min at room temperature in the dark.
-
Quantify the amount of protein in each sample using a protein assay (e.g. BSA tryptophan assay or Bradford assay)
Typically, 2×106 PMNs will yield 150 – 350 μg of total protein.
-
Add 1:50 (enzyme volume (μl):protein amount (μg)) of Lys-C protease (0.5 μg/μl) on ice.
For STAGE-tip purification (Basic Protocol 4, step 4), 50 μg of total protein is used; we recommend digesting 100 μg of total protein to use 50 μg for STAGE-tipping and 50 μg for short-term storage (up to 1 week) at −20°C with flash freezing in liquid nitrogen. Any remaining undigested total protein can be flash frozen in liquid nitrogen and stored at −20°C for up to 1 week or −80°C until needed.
Tap tube gently to mix.
-
Incubate at room temperature for 1 to 3 h.
For optimal digestion, we recommend incubation for 3 h.
Add 1:50 (enzyme volume (μl):protein amount (μg)) of trypsin protease (0.5 μg/μl) on ice.
Table tube gently to mix.
Incubate overnight at room temperature.
Stop the digestion by adding 1/10 volume stopping solution.
Centrifuge samples for 5 min at 10,000 × g in a bench-top centrifuge at room temperature to pellet any precipitate.
-
Transfer supernatants to new microcentrifuge tubes and discard the pellets.
At this point, the tube contains 100 ug of digested total protein, which can be used immediately for the STAGE-tip step (Basic Protocol 4, step 1) or can be flash frozen in liquid nitrogen and stored at −20°C for up to 1 week.
BASIC PROTOCOL 4
Proteomic profiling of neutrophils
This protocol describes profiling of the neutrophil proteome using a bottom-up mass spectrometry-based approach. Bottom-up proteomics relies on enzymatic digestion of proteins to peptides, followed by desalting and purification using STop And Go Extraction (STAGE) tips12, prior to separation by single-dimension C18 reverse phase nano-flow liquid chromatography couple to a tandem mass spectrometer. Samples are run over a 150 min chromatographic gradient with the mass spectrometer operating in data dependent mode for acquisition of mass-to-charge (m/z) profiles. The resulting mass spectrometry data are analyzed using MaxQuant with Andromeda3,8 for protein identification and quantification using label-free quantification9. Data interpretation and visualization if performed in Perseus4.
Materials:
Digested protein (Basic Protocol 3, step 17)
C18 resin (e.g. 3M Empore, cat no. 3M2215)
Buffer A (See Reagents and Solutions)
Buffer B (See Reagents and Solutions)
STAGE-tipping centrifuge (Sonation, cat no. STC-V2) or comparable
Vacufuge concentrator (e.g. Eppendorf, cat no. 07-748-15) or comparable
0.2 ml PCR tubes (e.g. ThermoFisher Scientific, cat no. AB0620) or comparable
Nanodrop (e.g. ThermoFisher Scientific, cat no. ND-2000) or comparable
Easy-Spray column, 50 cm × 75 μm, PepMap C18, 2 μm (e.g. ThermoFisher Scientific, cat no. ES803)
EASYnLC-1200 system (ThermoFisher Scientific, cat no. LC140) or comparable
QExactive quadrupole orbitrap mass spectrometer (ThermoFisher Scientific, cat no. 0726042) or comparable
Prepare C18 STAGE-tip by washing with 100 μl of 100% acetonitrile, centrifuge at 1,000 × g for 2 min.
Equilibrate C18 STAGE-tip by washing with 50 μl of Buffer B, spin at 1,000 × g for 2 min.
Equilibrate C18 STAGE-tip by washing with 200 μl of Buffer A, spin at 1,000 × g for 3 to 5 min.
-
Load approximately 50 μg of digested protein (from Basic Protocol 3, step 17) onto the C18 STAGE-tip, spin at 1,000 × g for 3 to 5 min.
Depending on the sample composition, a higher speed or longer spin times may be required to pass the sample through the C18 STAGE-tip. We recommend a maximum speed of 3,500 x g. Samples with high lipid composition or carry-over precipitate may require spin times of 20 min or longer.
-
Wash C18 STAGE-tip with 200 μl of Buffer A, spin at 1,000 × g for 3 to 5 min.
Again, depending on the ease or difficulty of spinning the sample through the C18 STAGE- tip, a higher spin rate or longer spin times, may be required for washing.
-
Elute peptides from C18 STAGE-tip with 50 μl of Buffer B, spin at 500 × g for 2 min into 0.2 ml PCR tubes.
For multiple samples, we recommend using 0.2 ml PCR strip tubes or a 96-well plate for elution.
-
Evaporate Buffer B from the sample using a Vacufuge concentrator at maximum speed for 30 to 40 min.
This step can be done at room temperature or 37°C for faster evaporation.
-
Resuspend peptides in 10 μl of Buffer A by pipetting up and down 3 to 5 times.
Samples can be loaded onto the mass spectrometer or stored at −20°C until needed.
-
Measure peptide concentration on Nanodrop to determine the volume needed to inject approximately 1.5 μg to 3.0 μg of peptides onto the mass spectrometry column.
Samples typically range from 0.6 to 1 μg/μl at a wavelength of A280 nm and sample type “1 Abs = 1 mg/ml”.
Inject the peptides into the mass spectrometer by separating over a 140 min gradient (5% to 60% acetonitrile in 0.5% acetic acid) followed by washout with up to 95% acetonitrile over a 10 min using a 50 cm Easy-Spray column heated to 50°C with a flow rate of 300 nl/min on the Easy-nLC 1200 system.
Operate a QExactive quadrupole orbitrap mass spectrometer in Top-15 data-dependent acquisition mode with full scans (m/z 300–1650) acquired in the Orbitrap analyzer with a resolution of 60,000 at 100 m/z.
-
Process the .RAW data files using MaxQuant software.
Filter protein identifications using a target-decoy approach at a false discovery rate of 1% with a minimum of two unique peptides for protein identification. Enable relative label-free quantification and match between runs, if desired. Using the Andromeda search engine (see Internet Resources section below), MaxQuant identifies and quantifies peptides present in a sample to proteins available in a user-inputted FASTA file for the relevant organism. User and experiment specific parameters can be set in consultation with available MaxQuant online resources (see Internet Resources section below).
-
Analyze the MaxQuant output file ‘proteingroups.txt’ in Perseus software for bioinformatic processing and visualization.
Analysis of MaxQuant output files in Perseus is user and experiment specific and can be performed in consultation with available Perseus online resources (see Internet Resources section below).
-
Deposit .RAW files and affiliated files into the ProteomeXchange consortium (PRIDE).
The PRIDE (PRoteomics IDEntification) database is a centralized, standards compliant, public data repository for proteomics data.
REAGENTS AND SOLUTIONS:
Buffer A
2% acetonitrile, 0.1% trifluoroacetic acid, 0.5% acetic acid, MS-grade water
Buffer B
80% acetonitrile, 0.5% acetic acid, MS-grade water
Phosphate buffer saline (PBS), pH 7.2 with 0.5% bovine serum albumin (BSA), 2 mM EDTA
PBS containing 0.5% BSA can be stored for up to 1 month at 4◦C, but will need to be sterile filtered, and 2 mM EDTA can be added when needed. It is not recommended to store buffer with EDTA at 4°C for long periods of time.
Flow cytometry buffer
PBS, 5% FBS
The solution needs to be 0.22 μM filter sterilized and used immediately. Fresh solution is made each time.
Neutrophil isolation media
25 ml PBS, 0.5% BSA, 2 mM EDTA
COMMENTARY:
BACKGROUND INFORMATION
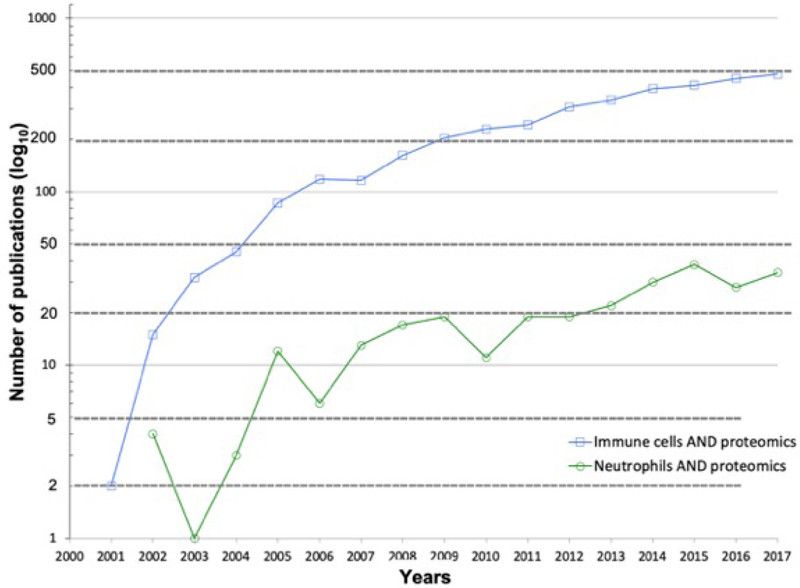
Over the past two decades, the field of immunology has benefited substantially from technological advances in mass spectrometry-based proteomics. Specifically, the application of mass spectrometry-based proteomics in the fields of innate immunity and neutrophil biology has resulted in a steady increase in contributions, as demonstrated in a search of PubMed publications (Figure 3). Proteomics based on high-resolution mass spectrometry is a powerful tool for profiling and quantifying proteins within cells, organs, or tissues1,2. It provides comprehensive information on the dynamics of cellular processes, modifications, and interactions. Discovery-driven proteomics is traditionally referred to as bottom-up or shotgun proteins, which relies on enzymatic digestion of proteins prior to identification on a mass spectrometer. Conversely, top-down proteomics encompasses the analysis of intact proteins and identification of proteoforms10, whereas targeted proteomics focuses on a limited set of predefined peptides in a a complex mixture and is often associated with biomarker development11.
Figure 3: Publications from 2000–2017 of proteomic applications in innate immunity.
An in-house developed R-script was used to search PubMed for publications using the following search terms within the abstract and/or title: i) “innate immune cells OR innate immunity AND proteomics”; and ii) “polymorphonuclear neutrophils OR neutrophils AND proteomics”. Note: y-axis plotted on log10 scale for improved data visualization.
For the purposes of this Protocol, we focus on bottom-up proteomics, which begins with sample preparation. In our experience, innate immune cells (e.g., neutrophils and macrophages) require mild disruption procedures, including chemical (e.g., urea) and mechanical (e.g., water bath sonication) approaches to sufficiently lyse the cells. Alternative and harsher disruption techniques may include extensive mechanical disruption (e.g., probe sonication) and boiling in a detergent (e.g., sodium dodecyl sulphate). The extracted proteins are then digested into peptides using sequence-specific proteases (e.g., Lys-C and trypsin) and purified on C18 STop And Go Extraction (STAGE)-tips12. For the absolute and relative quantification of proteins or peptides, metabolic (e.g. isotopically stable amino acid incorporation at the cellular level13), chemical (e.g., addition of mass tags or chemical derivatization14,15), or label-free9 (used in this Protocol) approaches are applied. In addition, samples may undergo separation based on size, mass, or charge to reduce sample complexity and promote deeper proteome coverage. Next, the first MS scan (MS1) records masses present at a given time and the subsequent MS2 or MS/MS scan selects and fragments peptides for identification based on fragment masses. As outlined in this Protocol, we employed a data-independent acquisition (DIA) strategy, where the top 15 most abundant ions are selected for fragmentation. An alternative approach, data-independent acquisition fragments all ions defined within a mass-to-charge (m/z) window, which is rapidly moved over the entire m/z range. Finally, the processing of modern MS data requires sophisticated bioinformatic workflows and platforms for data analysis, visualization, and interpretation. Here, we outline our use of the publicly-available and user-friendly platforms, MaxQuant3 and Perseus4. Lastly, to promote the development of machine learning tools (e.g., Big Data Analytics) and data sharing, we recommend deposition of data files and processing parameters into public repositories (e.g., The ProteomeXchange Consortium).
Neutrophils, derived from precursors in bone marrow, represent the most abundant leukocytes in blood and are indispensable in the host defense response to invading microbes16. The formation of granules by neutrophils provides protection of the host through exocytosis and/or exposure of soluble and membrane-bound proteins essential for neutrophil-endothelial interaction, extravasation, phagocytosis, and elimination of microbial cells. Moreover, the release of matrix-degrading enzymes from neutrophil storage granules during sample processing for proteomics experiments presents a challenge for comprehensive proteome profiling prior to degradation5,17. A seminal study of the neutrophil proteome, correlated proteomic and transcriptomic datasets, to reveal protein distribution profiles in granule subsets, secreted vesicles, and the surface membrane18. This study applied an in-gel digestion strategy to identify 1,292 known and previously uncharacterized neutrophil-associated proteins. Today, due to technological advances in mass spectrometry instrumentation, sample processing protocols, and bioinformatic platforms, proteome profiling of bone marrow-derived neutrophils from murine models at baseline and during infection by Pseudomonas aeruginosa at the ocular surface identified almost 4,300 neutrophil-associated proteins7. Here, exploration of the proteins displaying altered changes in abundance under the different conditions, suggested a novel strategy to combat biofilm formation at the ocular surface through the topical application of mannosidase. Taken together, defining the neutrophil proteome can provide valuable insight into the role of neutrophils during clearance of foreign invaders and mechanisms employed by the innate immune system to combat infection.
CRITICAL PARAMETERS AND TROUBLESHOOTING
For isolation of neutrophils, a single 6- to 8-week-old mouse yields approximately 40–50×106 cells after flushing the bones, with neutrophil yields in the range of 5–7×106 PMNs per mouse. These preparations are 80% enriched for murine PMNs. StemCell provides an alternative to the Biolegend PMN enrichment kit, which in our hands works comparably well. Both kits are based on a similar approach whereby, the purification of the neutrophils is based on depletion of the other bone marrow cells via biotinylated antibodies including anti-CD4, -CD5, -B220, - CD11c, -CX3CR1, -F4/80, -CD117, -CD244.2, and -TER-119 followed by streptavidin-bead-based capture. Cell viability should be estimated by standard approaches such as 7-AAD or PI staining by flow cytometry. Typically, the purification yields more than 70–80% viable cells. It is advisable to use polypropylene tubes when handling neutrophils as they become activated in the polystyrene tubes. For mass spectrometry-based proteomics analysis, gloves and lab coats should be worn at all times to limit contamination with keratins (e.g. hair and skin cells), which may interfere with LC-MS/MS analysis. In addition, work surfaces should be cleaned with 70% Ethanol prior to commencing the experiments to prevent sample contamination with dust. In the event of protein degradation or presence of a highly-abundant contaminant in the LC-MS/MS analysis, protein yields may be reduced. To improve protein identification rates, follow the notes listed above and increase the LC gradient length to reduce sample complexity. Table 2 describes common problems encountered with preparing proteomic samples, along with possible causes and recommended approaches to avoid or overcome these problems.
Table 2.
Troubleshooting Guide for Proteome Profiling of Neutrophils
| Problem | Possible cause | Solution |
|---|---|---|
| Low neutrophil enrichment yields | Inefficient negative selection | Add an additional magnet capture step to the purification protocol |
| Low viability of PMNs <75% | Extended purification time or Erythrocyte lysis | Avoid red cell lysis |
| Poor digestion of peptides | Concentration of Urea is too high during trypsin digestion | Ensure that urea is at 2 to 2.5 M during trypsin digestion; check pH during digestion (pH 8 is optimal) |
| Lys-C and trypsin proteases are not working optimally | Use fresh proteases stored according to manufacture’s instructions. Keep proteases on ice at all times during use and discard any remaining protease after thawing. | |
| Poor peptide yield | In-house STAGE-tips were packed too tightly/loosely | Ensure recommended centrifugation speeds and times are accurate for Buffer A; if tip is packed too loosely, Buffer A will pass very quickly and if packed too tightly, higher speeds and time will be required to wash the tip |
| STAGE-tip is not sufficiently activated or has run dry during loading | Ensure that C18 resin never runs dry during activation, loading, or washing |
ANTICIPATED RESULTS:
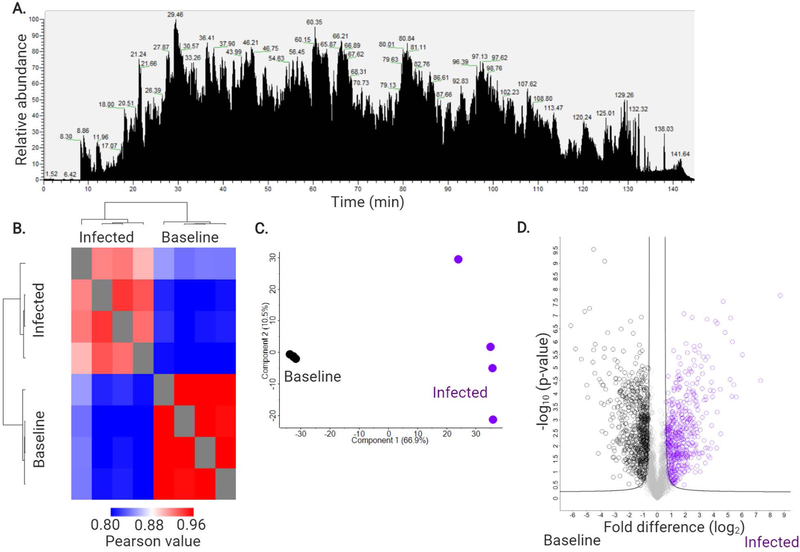
Using the protocols described above, we have routinely obtained 80% enrichment for murine neutrophils with yields of 5–7 × 106 cells per mouse. Depending on the quantity and quality of the starting material, and the proteomics technologies available (e.g. mass spectrometer instrumentation), >3,500 proteins should be identified from the neutrophil samples (Figure 4).
Figure 4: Expected MS results.
A) An example mass spectrometry chromatogram of peptides extracted from purified neutrophils of C57BL6/N mice at baseline ran on 150 min gradient on QExactive HF quadrupole orbitrap mass spectrometer following reverse-phase separation by Easy1200-nLC system. Intensity = 1.98E10. B) Heatmap of Pearson correlation followed by hierarchical clustering by Euclidean distance to demonstrate condition separation and quantify replicate reproducibility (i.e. 94.0% C57BL6/N at baseline and 94.1% at infection). C) Principal component analysis demonstrating separation between C57BL6/N baseline (dark grey) and infected (purple) neutrophils. Four biological replicates were processed per condition. D) Volcano plot of all proteins identified in the analysis. Significantly different proteins with an increase in abundance during C57BL6/N infection are highlighted in purple; significant increase in abundance at C57BL6/N baseline are highlighted in dark grey. Student’s t-test p-value < 0.05; FDR = 0.01; S0 = 1.
TIME CONSIDERATIONS:
Purification of neutrophils from murine bone marrow takes 1 h for an average of 4 mice but can vary depending on the number of mice. Flow cytometry should be performed on the samples immediately following purification and requires approximately 1.5 h. Protein extraction, including sonication, reduction, alkylation, and quantification takes approximately 2 h and is followed by enzyme digestion of proteins into peptides, requiring an overnight incubation (1 day). Desalting and purification of the peptides by STAGE-tip takes approximately 3 h, including tip activation, washing, sample loading, eluting, and drying. Samples can be immediately measured on the mass spectrometer; each sample may take approximately 3 h to analyze using a 150 min gradient (i.e. 10 samples would take 1.5 days to measure). Database searching can typically be performed in 1 day with subsequent statistical processing and comprehensive analysis of the datasets taking 3 to 4 days. In total, this Protocol requires approximately 7 to 10 days of dedicated time to be performed.
SIGNIFICANCE STATEMENT:
Over the past two decades, the field of immunology has benefited substantially from technological advances in mass spectrometry-based proteomics. Specifically, MS-based proteomics provides a robust and sensitive platform for profiling and quantifying protein content of cells, organs, or tissues. It provides comprehensive information about the dynamics of cellular processes, modifications, and interactions. For the innate immune system, proteomic profiling defines communication patterns among immune cells and provides insights into differences in patterns of regulation during health and disease. With this information, we can identify specific proteins or pathways critical for effective host immune response and suggest novel therapeutic strategies to combat a diverse array of diseases.
ACKNOWLEDGEMENTS:
This work was supported, in part, by the Natural Sciences and Engineering Research Council of Canada (NSERC) and the Alexander von Humboldt Foundation for J.G.-M. and by the NIH/NEI EY022054 for M.G.
LITERATURE CITED:
- 1.Aebersold R & Mann M Mass-spectrometric exploration of proteome structure and function. Nature 537, 347–355 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Mann M, Kulak NA, Nagaraj N & Cox J The Coming Age of Complete, Accurate, and Ubiquitous Proteomes. Mol. Cell 49, 583–590 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Cox J & Mann M MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Tyanova S et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature Methods 13, 731–740 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Ley K, Laudanna C, Cybulsky MI & Nourshargh S Getting to the site of inflammation: The leukocyte adhesion cascade updated. Nature Reviews Immunology (2007). doi: 10.1038/nri2156 [DOI] [PubMed] [Google Scholar]
- 6.Kugadas A, Wright Q, Geddes-McAlister J & Gadjeva M Role of microbiota in strengthening ocular mucosal barrier function through secretory IgA. Investig. Ophthalmol. Vis. Sci (2017). doi: 10.1167/iovs.17-22119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kugadas A et al. Employing enzymatic treatment options for management of ocular biofilm-based infections. J. Leukoc. Biol (2019). doi: 10.1002/JLB.4HI0918-364RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cox J et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. (2011). doi: 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- 9.Cox J et al. Accurate Proteome-wide Label-free Quantification by Delayed Normalization and Maximal Peptide Ratio Extraction, Termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toby TK, Fornelli L & Kelleher NL Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. (Palo Alto. Calif) (2016). doi: 10.1146/annurev-anchem-071015-041550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aebersold R, Bensimon A, Collins BC, Ludwig C & Sabido E Applications and Developments in Targeted Proteomics: From SRM to DIA/SWATH. Proteomics (2016). doi: 10.1002/pmic.201600203 [DOI] [PubMed] [Google Scholar]
- 12.Rappsilber J, Mann M & Ishihama Y Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc 2, 1896–1906 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Ong S-E et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc. Natl. Acad. Sci 106, 4617–4622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter SV et al. EASI-tag enables accurate multiplexed and interference-free MS2-based proteome quantification. Nat. Methods (2018). doi: 10.1038/s41592-018-0037-8 [DOI] [PubMed] [Google Scholar]
- 15.Thompson A et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem 75, 1895–1904 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Kolaczkowska E & Kubes P Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol (2013). doi: 10.1038/nri3399 [DOI] [PubMed] [Google Scholar]
- 17.Luerman GC, Uriarte SM, Rane MJ & McLeish KR Application of proteomics to neutrophil biology. Journal of Proteomics (2010). doi: 10.1016/j.jprot.2009.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rørvig S, Østergaard O, Heegaard NHH & Borregaard N Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J. Leukoc. Biol (2013). doi: 10.1189/jlb.1212619 [DOI] [PubMed] [Google Scholar]
INTERNET RESOURCES:
MaxQuant
MaxQuant is a quantitative proteomics software package designed for analyzing large mass spectrometric data sets. MaxQuant is freely available and can be downloaded from the provided link. Wiki documentation is also provided and updated regularly to assist users and an annual Summer School provides hands-on training.
http://www.coxdocs.org/doku.php?id=maxquant:start
https://maxquant.org/summer_school/
Andromeda
A peptide search engine based on probabilistic scoring (comparable to the commercially-available Mascot) with independent and integrated functionality with MaxQuant.
http://www.coxdocs.org/doku.php?id=maxquant:andromeda
Perseus
The Perseus software platform supports biological and biomedical researchers in interpreting protein quantification, interaction, and post-translational modification data. It provides a user-friendly, interactive workflow environment with complete documentation of computational methods used for publication. Wiki documentation is also available and updated regularly to assist users.
http://www.coxdocs.org/doku.php?id=perseus:start
PRIDE
https://www.ebi.ac.uk/pride/archive/
The PRIDE (Proteomics IDEntification) database is a centralized, standards compliant, public data repository for proteomics data, including protein and peptide identifications, post-translational modifications and supporting spectral evidence.