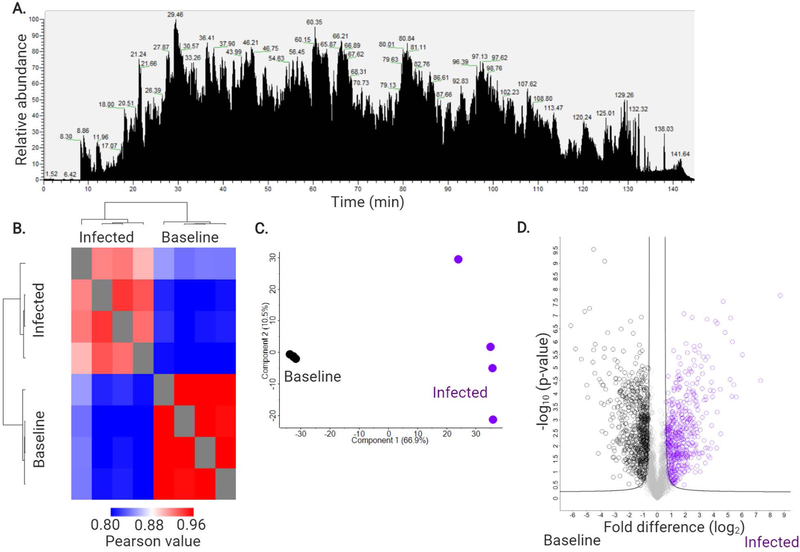
Figure 4: Expected MS results.
A) An example mass spectrometry chromatogram of peptides extracted from purified neutrophils of C57BL6/N mice at baseline ran on 150 min gradient on QExactive HF quadrupole orbitrap mass spectrometer following reverse-phase separation by Easy1200-nLC system. Intensity = 1.98E10. B) Heatmap of Pearson correlation followed by hierarchical clustering by Euclidean distance to demonstrate condition separation and quantify replicate reproducibility (i.e. 94.0% C57BL6/N at baseline and 94.1% at infection). C) Principal component analysis demonstrating separation between C57BL6/N baseline (dark grey) and infected (purple) neutrophils. Four biological replicates were processed per condition. D) Volcano plot of all proteins identified in the analysis. Significantly different proteins with an increase in abundance during C57BL6/N infection are highlighted in purple; significant increase in abundance at C57BL6/N baseline are highlighted in dark grey. Student’s t-test p-value < 0.05; FDR = 0.01; S0 = 1.