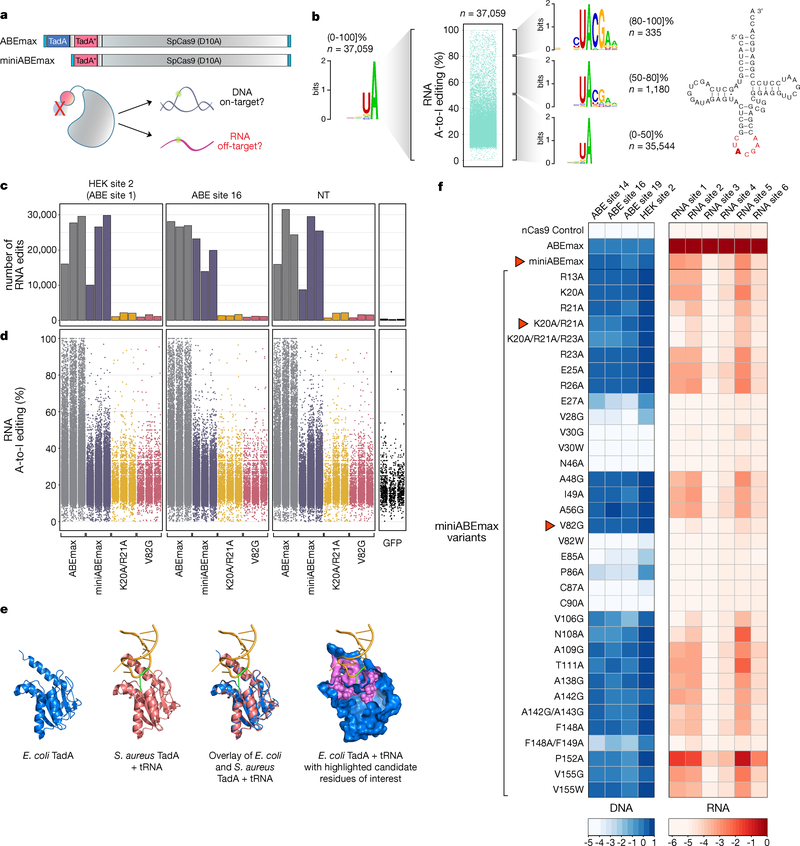
Figure 1. Engineering of SECURE-ABE variants with reduced off-target RNA editing activities.
(a) Schematic illustration of ABEmax and miniABEmax architectures and overview of experimental testing of miniABEmax for on-target DNA and off-target RNA editing. Light blue boxes = bipartite NLS at N- and C-termini, TadA* = mutant TadA 7.103, and small grey boxes = 32AA flanked XTEN linkers. nCas9 (SpCas9 D10A) = grey shape, TadA WT and mutant monomers = blue and red circles. Green halo = sites of potential adenine deamination on DNA and RNA. (b) Unstratified sequence logo (left) and stratified sequence logos for RNA adenines edited with high (80–100]%, middle (50–80]%, and low (0–50]% efficiencies (right) by ABEmax. n= number of modified adenines. RNA-seq data shown in the Jitter plot was obtained from HEK293T cells in an earlier published study5. Cloverleaf structure of E. coli tRNAArg2 (Ref. 8), illustration adapted from Fig. 1a of Ref. 11. Anticodon loop highlighted in red (Ref. 8). TadA target adenine 34 is highlighted in bold. (c) Bar plots showing the number of RNA A-to-I edits observed in RNA-seq experiments in HEK293T cells with expression of ABEmax, miniABEmax, miniABEmax-K20A/R21A, or miniABEmax-V82G each with three different gRNAs (HEK site 2, ABE site 16, and non-targeting (NT)) and performed in independent replicates (n = 3). Exact number of edits are shown in Supplementary Table 1. GFP negative controls performed as independent replicates (n = 3) are also shown. (d) Jitter plots showing the efficiencies of RNA A-to-I edits from the RNA-seq experiments shown in c. Each dot represents an edited adenine position in RNA. (e) Structural representations of E. coli TadA (PDB 1Z3A), structural representation of S. aureus TadA in complex with tRNA (PDB 2B3J), overlaid structures from E. coli TadA and S. aureus TadA, and surface representation of E. coli TadA in blue with backbone carbons of amino acid positions proximal to the predicted deaminase catalytic site highlighted in pink. Target adenine on tRNA (A34) marked in green. All graphical representations generated with PyMol (Methods). (f) Testing of 34 miniABEmax variants for their on-target DNA editing (A-to-G) and off-target RNA editing (A-to-I) activities. On-target DNA editing was assessed with four different gRNAs and off-target RNA alterations were screened on six RNA adenines previously identified as being efficiently modified by ABEmax5. Efficiencies are shown in heat map format (log2-fold changes), with each box representing the mean of four independent replicates normalized to the editing efficiency observed with ABEmax for each target DNA or RNA off-target site. Red arrows indicate three variants that were chosen for further analysis. Amino acid abbreviations are according to IUPAC nomenclature and residue numbering is based on the amino acid position in E. coli TadA. A = adenine; I = inosine. ABEmax = codon optimized adenine base editor. miniABEmax = ABEmax without N-terminal wild type TadA domain and the proximal 32AA linker.