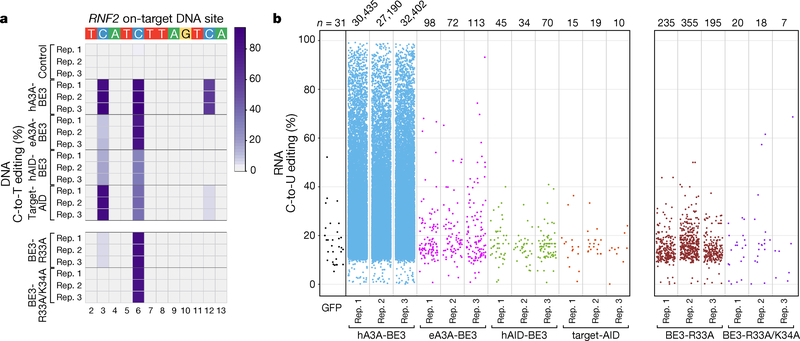
Figure 3. Transcriptome-wide off-target RNA editing activities of CBEs with non-APOBEC1 cytidine deaminases in HEK293T cells.
(a) Heat map showing the on-target DNA editing efficiencies of nCas9-UGI (Control), hA3A-BE3, eA3A-BE3, hAID-BE3, Target-AID, SECURE BE3-R33A and BE3-R33A/K34A with a gRNA targeted to the RNF2 gene (n= 3 independent replicates). Editing window shown includes only the most highly edited cytosines and not the entire spacer sequence. Numbering at the bottom represents spacer position with 1 being the most PAM distal location. (b) Jitter plots showing transcriptome-wide RNA C-to-U edits observed with a GFP negative control (single replicate) and hA3A-BE3, eA3A-BE3, hAID-BE3, Target-AID, SECURE BE3-R33A and BE3-R33A/K34A (each n= 3 independent replicates). Each dot represents a single edited cytosine. All experiments (except for the GFP control) were performed with co-expression of a gRNA targeting a site in the RNF2 gene and in all experiments the cells were sorted for top 5% of GFP signal except for the GFP control which was sorted for equivalent MFI of top 5% BE3 (Methods)). All CBEs (except Target-AID) used nCas9-UGI as negative control for RNA variant calling; Target-AID used NLS-nCas9-NLS-SH3-UGI (Target-AID without pmCDA1) as a negative control (Supplementary Table 3; Methods). n = total number of modified cytosines. DNA on-target data and RNA-seq data for BE3-R33A and BE3-R33A/K34A presented are from an earlier published study5 (performed using the same experimental conditions) and are shown here to facilitate direct comparison with other CBEs.