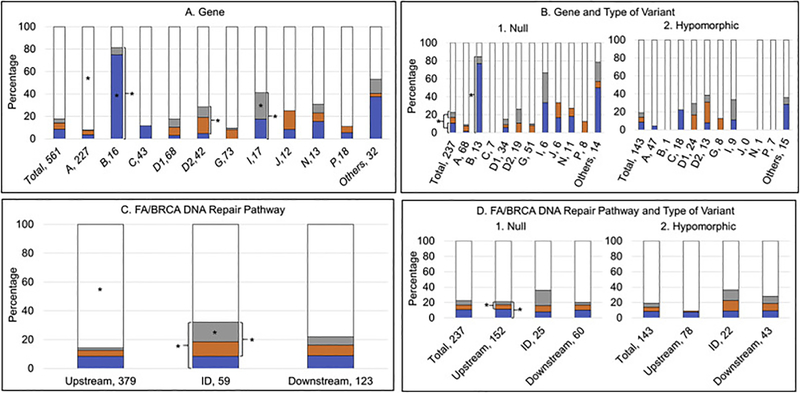
Fig. 6.
Presence or absence of VACTERL-H and/or PHENOS. Blue, VACTERL-H alone; orange, PHENOS alone; gray, VACTERL-H plus PHENOS; white, neither VACTERL-H nor PHENOS. A and B) Horizontal axis: gene, number of patients; vertical axis: percent of cases within each gene. C and D) Horizontal axis: location in the pathway, number of patients; vertical axis: percent of cases within each location in the pathway. *p<0.05. (A) 9% of all patients had VACTERL-H alone, 5% had PHENOS alone, 4% had VACTERL-H plus PHENOS, and 82% had neither VACTERL-H nor PHENOS. A higher proportion of patients with FANCB and I had VACTERL-H (irrespective of PHENOS) (p<0.001 and p=0.02, respectively). A higher proportion of patients with FANCD2 and I had PHENOS (irrespective of VACTERL-H) (p=0.01 for FANCD2). A higher proportion of patients with FANCB had VACTERL-H alone (without PHENOS) (p<0.001). VACTERL-H plus PHENOS was more frequent in patients with FANCI (p=0.03). VACTERL-H and/or PHENOS was rare in patients with FANCA (p<0.0001). (B) Overall, VACTERL-H (irrespective of PHENOS) was more frequent in patients with biallelic or hemizygous null variants than in those with at least one hypomorphic allele (p=0.01). Gene by gene, VACTERL-H (irrespective of PHENOS) and at least one of the two (VACTERL-H or PHENOS) were more frequent in patients with biallelic or hemizygous null variants in FANCB (p<0.05) and I. (C) VACTERL-H and/or PHENOS were less frequent in patients with pathogenic variants in upstream genes (p<0.0001). A higher proportion of patients with pathogenic variants in ID complex genes had PHENOS (irrespective of VACTERL-H) (p=0.001), both VACTERL-H plus PHENOS (p=0.003), or at least one of the two (p=0.01) compared with patients with pathogenic variants in upstream and downstream genes. (D) Patients with biallelic or hemizygous null variants in upstream genes had a higher proportion of PHENOS (irrespective of VACTERL-H) and at least one of the two (VACTERL-H or PHENOS) than those with hypomorphic genotype (p<0.02); these associations were not seen in ID or downstream genes.