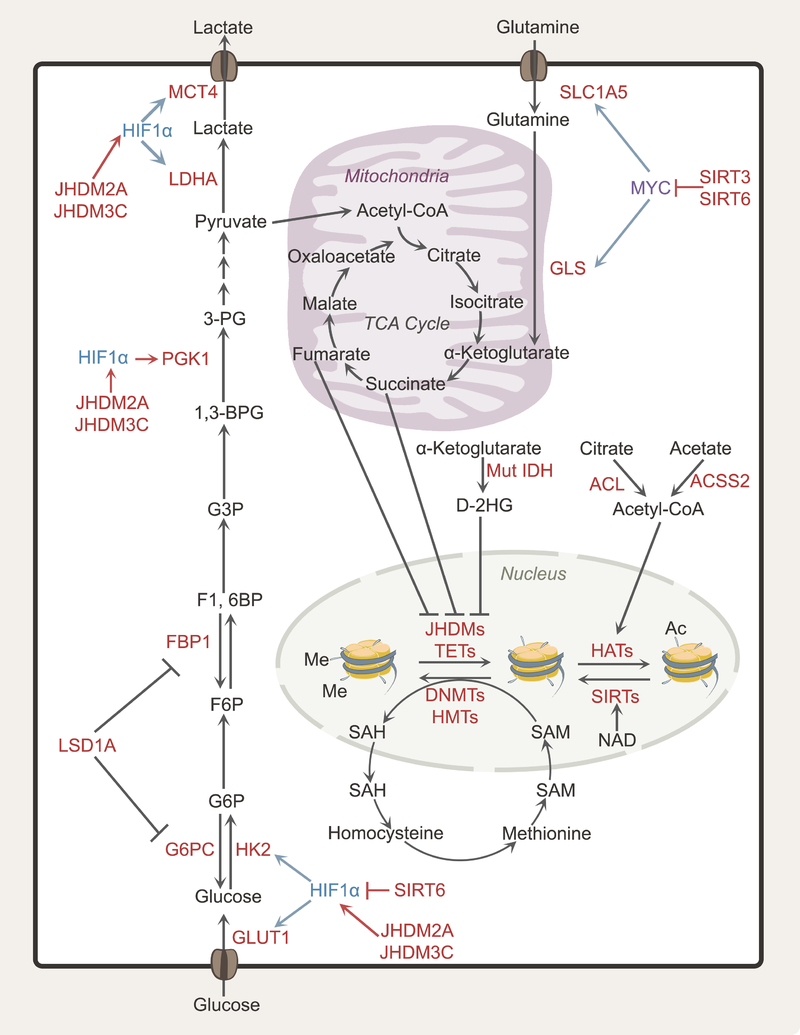
Figure 3. Epigenetics and metabolism in cancer cells.
Epigenetic regulation of gene expression contributes to metabolic heterogeneity because many metabolic enzymes and nutrient transporters are regulated by epigenetic modifications of histones (acetylation, methylation) and DNA (methylation). Conversely, these epigenetic modifications respond to the metabolic state of the cell. The relative abundances of SAM and SAH regulate DNA and histone methyltransferases, while the abundance of acetyl-CoA, the ratio of acetyl-CoA to free CoA and NAD levels can regulate histone acetylation. The TCA cycle intermediate α-KG affects demethylation of histones and DNA by acting as a co-substrate for JHDM histone demethylases and TET-family methylcytosine dioxygenases, respectively. The oncometabolites fumarate, succinate and 2-HG are dicarboxylates that compete with α-KG, interfering with TET/JHDM function. Abbreviations: HK2, hexokinase 2; MCT4, monocarboxylate transporter 4; SAM, S-adenosylmethionine; SAH, S-adenosyl-homocysteine; NAD, nicotinamide adenine dinucleotide; α-KG, alpha-ketoglutarate; 2-HG, 2-hydroxyglutarate; TET, 10–11 translocation enzyme; JHDM, Jumonji C domain-containing histone demethylase.