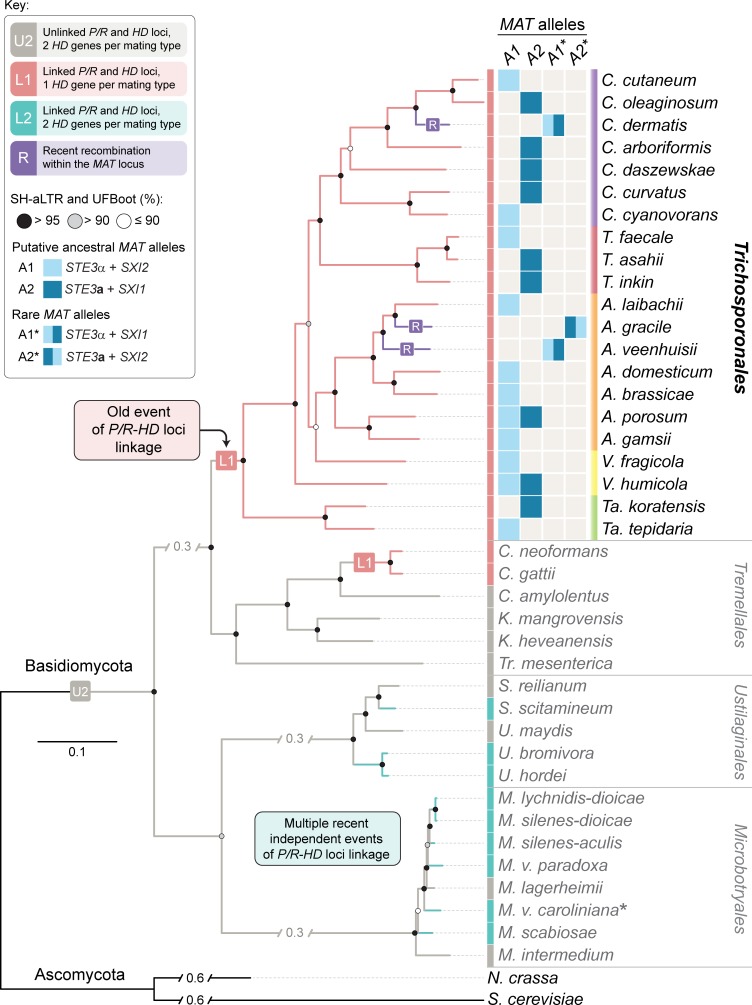
Fig 1. Overview of the haploid Trichosporonales species for which MAT regions were analyzed.
Shown here is a phylogenetic tree of basidiomycetes with a focus on groups where fusion of MAT loci occurred as was shown previously (Cryptococcus, Ustilago, Malassezia, Microbotryum, Sporisorium) [4, 8, 18]. The phylogenetic branching within the tree agrees with previous studies [4, 8, 26–28]. Next to each Trichosporonales species name (color coded according to their genus), the MAT alleles that are represented in the sequenced genomes are shown. Within one species, different strains carrying different alleles are currently known only for V. humicola and A. porosum. Abbreviations for Trichosporonales genus names are: A—Apiotrichum, C—Cutaneotrichosporon, T—Trichosporon, Ta—Takashimella, V—Vanrija. Abbreviations for other basidiomycete genus names are: C—Cryptococcus, K—Kwoniella, M—Microbotryum, S—Sporisorium, Tr—Tremella, U—Ustilago. Ascomycetes Neurospora crassa and Saccharomyces cerevisiae were used as outgroups. Branch lengths are given in number of substitutions per site (scale bar). Branch support was assessed by 10,000 replicates of both ultrafast bootstrap approximation (UFBoot) and the Shimodaira-Hasegawa approximate likelihood ratio test (SH-aLRT) and represented as circles colored as given in the key. The asterisk next to the species name for Microbotryum violaceum caroliniana indicates that the HD1 gene was specifically lost in the A2 mating type of this species [8].