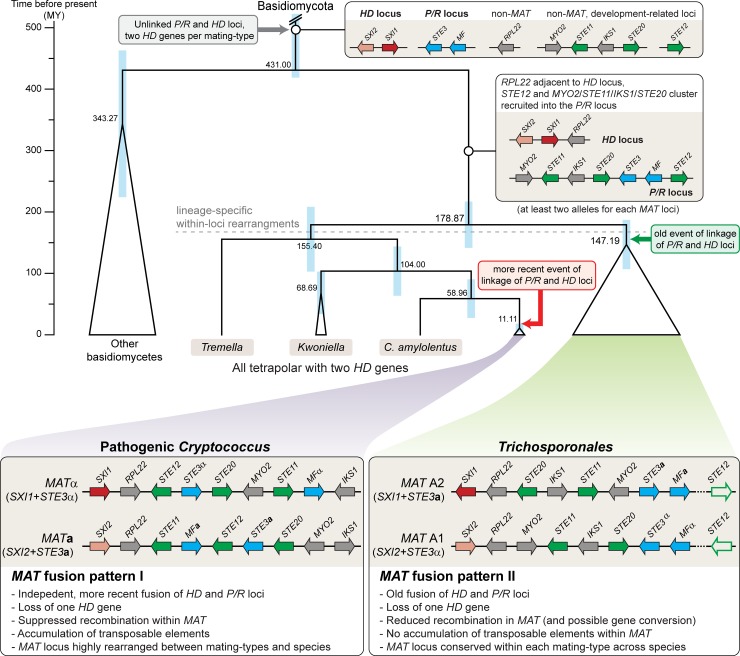
Fig 5. Model for the evolution of the MAT loci in Tremellomycetes.
Genes at the core MAT loci include the homeodomain transcription factor genes (HD locus, shown in red/pink) and the pheromone and receptor genes (P/R locus, shown in blue). Genes involved in sexual development, but not originally part of a MAT locus are shown in green, other genes are shown in grey. Only genes from the C. neoformans MAT locus that are also linked to the core MAT genes (STE3, HD genes) in the Trichosporonales are shown (STE11, STE12, STE20, IKS1, MYO2, RPL22). Other genes present at the MAT loci are left out for clarity. A trend towards integrating other developmental genes into the MAT loci is reflected in the recruitment of the STE12 gene and an ancestral STE11/STE20 cluster into the P/R locus. This resulted in a tetrapolar arrangement with (at least) two alleles for each MAT locus. In the Tremellales, tetrapolarity is retained in the extant Tremella, Kwoniella, and non-pathogenic Cryptococcus lineages, whereas in the pathogenic Cryptococci, the MAT loci fused and one HD gene was lost from each MAT allele. Fusion of the MAT loci occurred independently and earlier in the ancestor of the Trichosporonales, also with loss of one HD gene, resulting in two known MAT alleles with combinations STE3a-SXI1 and STE3α-SXI2 that differ from those in the known Cryptococcus alleles. The STE12 gene is shown in outline only to indicate that it is not part of the MAT locus in all species; STE12 may have been recruited to the P/R locus in the Tremellales-Trichosporonales common ancestor and eventually evicted from the MAT locus in the Trichosporonales possibly associated with the fusion event of P/R and HD loci, or instead STE12 was recruited to MAT only in the Tremellales. The phylogenetic relationships and inferred dates of speciation (numbers in black on tree nodes) are depicted according to S7 Fig. The blue bars correspond to 95% confidence intervals.