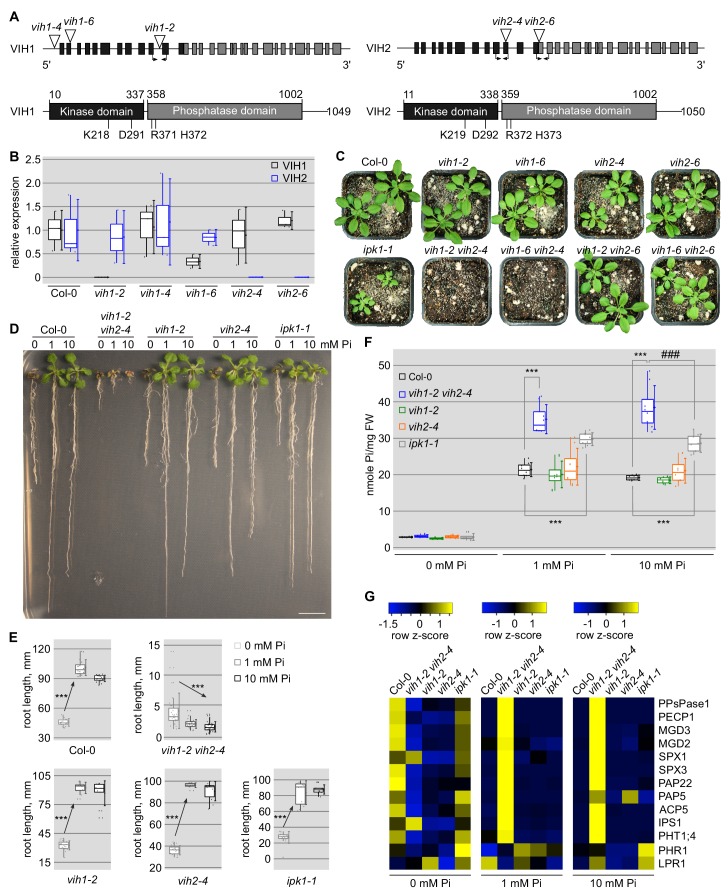
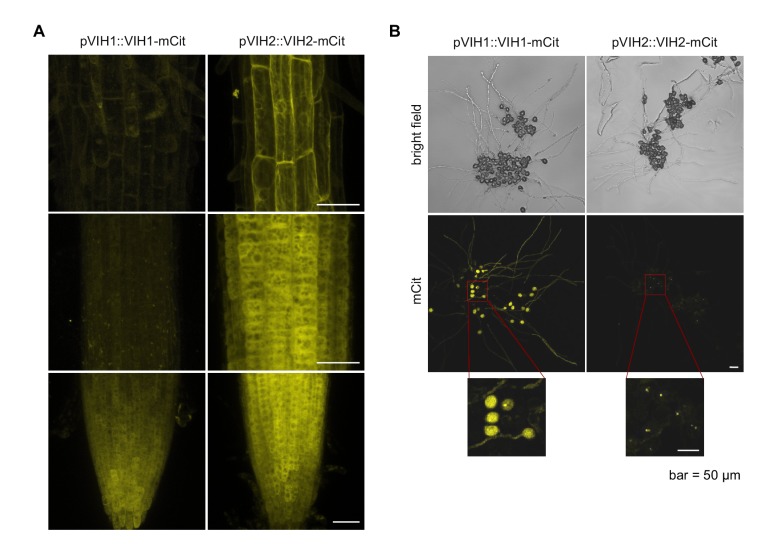
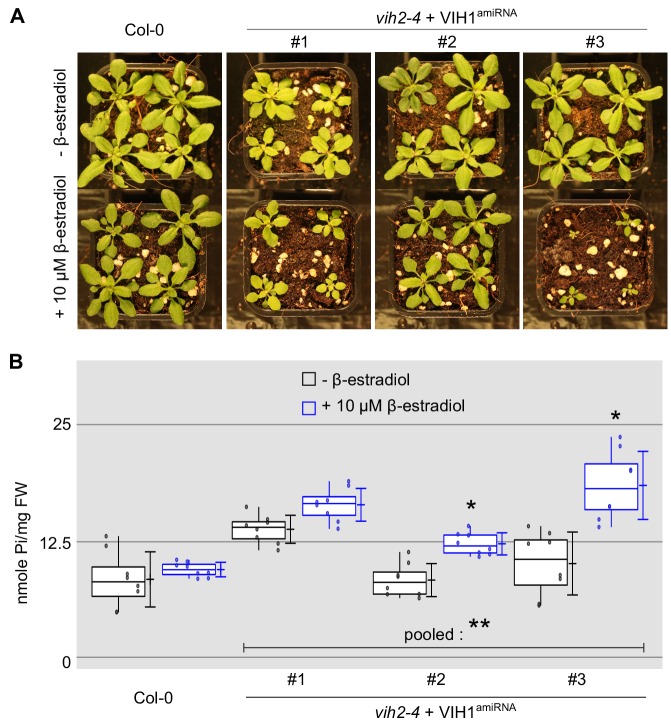
Figure 1. vih1 vih2 loss-of-function mutants show severe growth phenotypes and hyperaccumulate Pi.
(A) Schematic overview of VIH1 and VIH2: (upper panel) VIH1 and VIH2 genes with exons described as rectangles, introns as lines. T-DNA insertions are depicted as triangles, primer positions used in qRT-PCR analyses are indicated by arrows. (lower panel) VIH1 and VIH2 protein architecture, with kinase domains shown in black, phosphatase domains in gray and putative linkers/unstructured regions as lines. The point mutations used in this study are shown below the domain schemes. (B) qRT-PCR expression analysis of VIH1 and VIH2 transcripts in the T-DNA mutant allele backgrounds. Shown are 2-∆CT values relative to Col-0 wild-type. Quantifications were done using three biological replicates. (C) Growth phenotypes of vih1 and vih2 single mutant, and of vih1 vih2 double mutants. Shown are plants 20 DAG in soil compared to Col-0 wild-type. (D) Growth phenotypes of vih1-2 vih2-4, vih1-2, vih2-4, and of ipk1-1 compared to Col-0 wild-type seedlings. Plants were germinated in vertical 1/2MS plates for 8 d, transferred to Pi-deficient 1/2MS plates supplemented with either 0 mM, 1 mM or 10 mM Pi and grown for additional 6 d. Scale bars correspond to 2 cm. (E) Trend analysis of seedling root length vs. cellular Pi concentration for the seedlings described in (D). For each boxed position, root length measurements were performed for seedlings from three independent MS plates. (F) Pi contents of the seedlings shown in (D) 14 DAG. For each boxed position, four independent plants were measured with two technical replicates. (G) qRT-PCR quantification of PSI marker genes (PPsPase1, PECP1, MGD3, MGD2, SPX1, SPX3, PAP22, PAP5, ACP5, IPS1, PHT1;4) and of the non-PSI genes PHR1 and LPR1 in Col-0 wild-type, vih1-2 vih2-4, vih1-2, vih2-4 and ipk1-1 seedlings described in (D). Expression levels are represented as Z-scores. The original data are shown in Supplementary file 3a.