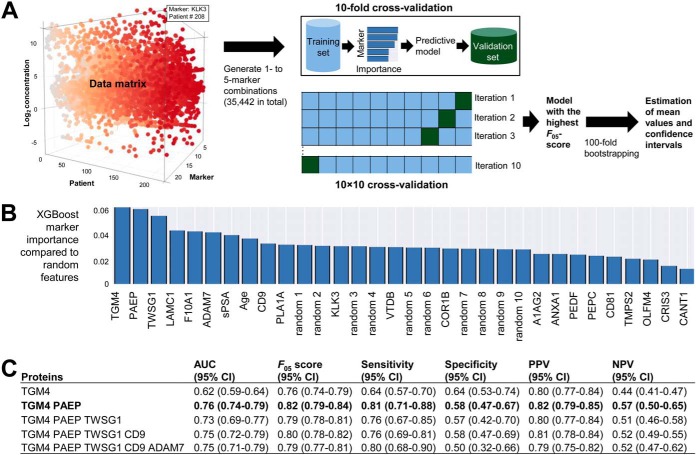
Fig. 4.
Machine-learning analysis to identify multi-variable combinations of markers. A, Twenty two variables (19 candidate SP proteins, seminal KLK3, serum PSA and age) were used to generate all possible 1- to 5- marker combinations. XGBoost algorithm was applied to identify combinations with the highest F05-measure scores and calculate AUCs, sensitivities, specificities, PPVs and NPVs. Stringent 10 × 10 cross-validation was applied to reduce over-fitting. Top combinations were verified on the whole dataset of patients to ensure that each potential marker had feature scores higher than a randomly generated feature. Finally, 100-fold bootstrapping was used to estimate mean values for performance metrics and calculate 95% confidence intervals. B, XGBoost importance of individual markers to differentiate between PCa and negative biopsy, as compared with random features. C, Diagnostic performance of top combinations, with 95% confidence intervals estimated using 100-fold bootstrapping. Combination of TGM4 with PAEP protein improved AUC and sensitivity to differentiate between negative biopsy and PCa, whereas additional markers did not further increase AUCs.