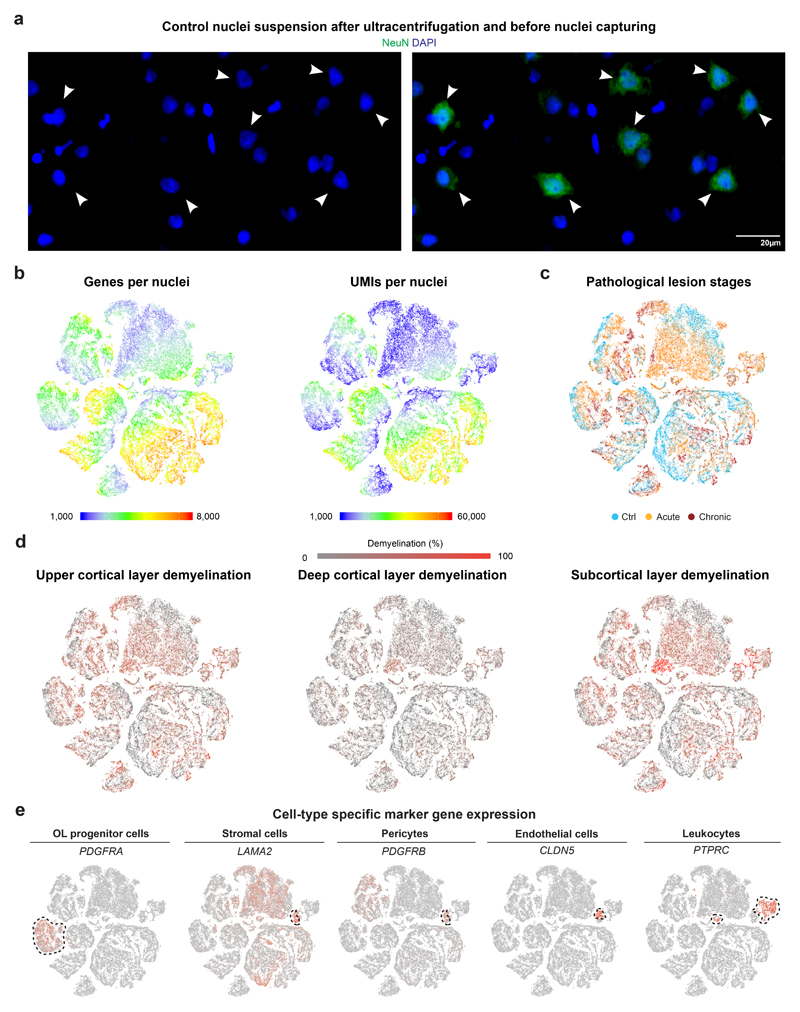
Extended Data Fig. 1. Sample and disease contribution of cell types captured by snRNA-seq.
(a) Representative images selected from nuclei suspensions (ctrl, n=9; MS, n=12) after ultracentrifugation and before capturing by 10X Genomics confirming DAPI nuclear counterstaining with presence of smaller and larger DAPI+ nuclei. Note that larger nuclei are co-stained with anti-NeuN antibody confirming neuronal origin (white arrowheads). (b) Colored t-SNE plots showing numbers of genes (left) and UMIs (right) per captured nuclei from control and MS samples. (c) Colored t-SNE plot visualizing nuclei from different lesion stages based on classic pathological MS lesion staging. (d) Colored t-SNE plots visualizing nuclei from samples with different levels of upper and deep layer cortical demyelination as well as subcortical demyelination. (e) Representative tSNE plots with cell-type specific marker genes for OL progenitor cells, stromal cells including pericytes, endothelial cells, and leukocytes. For tSNE plots, data shown from 9 control and 12 MS samples and a total of 48,919 nuclei.