Figure 5. Importance of ApaH and RppH for RNA decapping and degradation in E. coli.
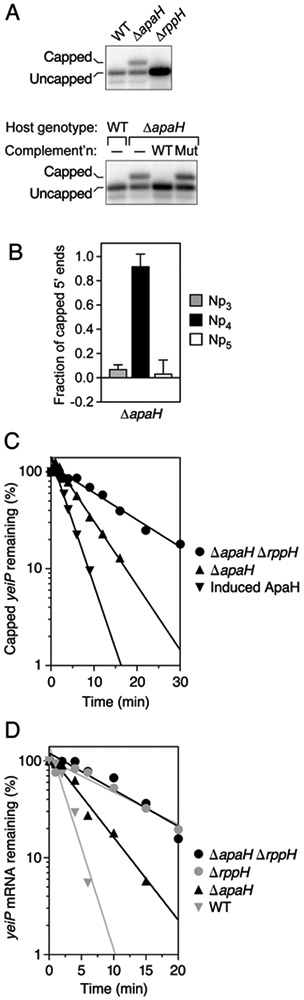
(A) Detection of capped yeiP mRNA in cells that lack ApaH but not in cells that lack RppH. Total RNA was extracted from unstressed cultures of isogenic wild-type, ΔapaH, or ΔrppH cells (top) or from ΔapaH cells complemented with a plasmid encoding wild-type or catalytically inactive ApaH (bottom), and the presence of capped yeiP mRNA was examined by boronate gel electrophoresis and blotting. WT, wild-type apaH allele; Mut, mutant apaH allele encoding a catalytically inactive enzyme; –, no plasmid. See also Fig. S2B.
(B) Calculated fraction of capped yeiP 5′ ends bearing an Np3, Np4, or Np5 cap in ΔapaH cells, as determined by esCAPade. The data from Fig. S7A and two additional biological replicates were used to calculate each value (Table S6). Error bars correspond to a confidence level of 68.3%. See also Figs. S1 and S7A and Tables S1-S6.
(C) Rate of loss of capped yeiP mRNA after arresting transcription in E. coli apaH and rppH mutants. Cultures of the indicated strains growing without stress in LB were treated with rifampicin to arrest transcription, and equal amounts of total RNA extracted at time intervals thereafter were analyzed by boronate gel electrophoresis and blotting. Data from representative experiments are shown. See also Fig. S2C.
(D) Effect of an apaH deletion on the aggregate decay rate of yeiP mRNA in the presence or absence of RppH. Cultures of the indicated strains growing without stress in MOPS-glucose were treated with rifampicin, and equal amounts of total RNA extracted at time intervals thereafter were analyzed by electrophoresis on ordinary polyacrylamide gels. Data from representative experiments are shown. See also Fig. S2D.
