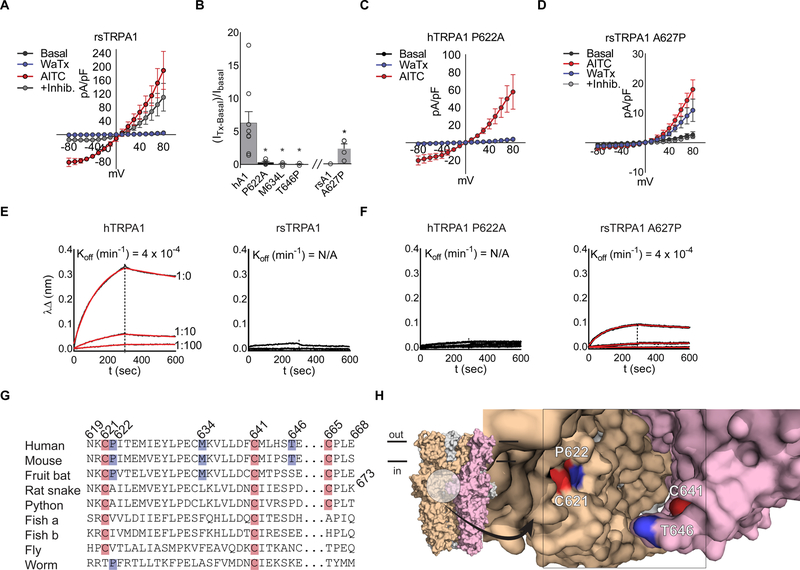
Figure 3. WaTx interacts directly with TRPA1’s allosteric nexus.
(A) Rat snake (rs) TRPA1 current-voltage relationships in transfected HEK cells. WaTx (100 nM), AITC (500 μM), AITC + inhibitor (50 μM, HC 030031); n = 4 cells. Note: inhibitor A 967079 is ineffective against rsTRPA1 while the selective TRPA1 inhibitor HC 030031 is effective (Banzawa et al., 2014; McNamara et al., 2007).
(B) WaTx-evoked whole cell currents for loss-of-function hTRPA1 mutants and gain-of-function rsTRPA1 mutant. WaTx treatments: 1 μM to hTRPA1 and 5 μM to rsTRPA1 constructs. One-Way ANOVA with post-hoc Holm-Sidak correction for multiple comparisons (hTRPA1 constructs) and unpaired two-tailed Student’s t-test (rsTRPA1 constructs.). Recordings from transfected HEK cells; n = 3–9 cells/construct.
(C, D) Current-voltage relationships for hTRPA1 P622 mutants, which dictate species selectivity of TRPA1. Recordings from transfected HEK cells; n = 4–5 cells/treatment.
(E, F) Biolayer-interferometry derived sensorgrams depicting association of crude HEK cell membranes expressing TRPA1 to WaTx-coated sensors. Non-specific binding to mock-transfected membranes was subtracted, and resultant curves fit by nonlinear regression to a one-phase exponential association and dissociation model; Data are representative of n = 3–6 independent experiments/construct.
(G) Multiple-sequence alignment of TRPA1 orthologs and (H) proposed WaTx binding pocket mapped to the cryo-EM structure of human TRPA1 (PDB ID: 3J9P). Electrophile-reactive cysteines highlighted red; residues implicated in WaTx binding, blue.
All summary data, mean ± SEM. See also Figure S3.