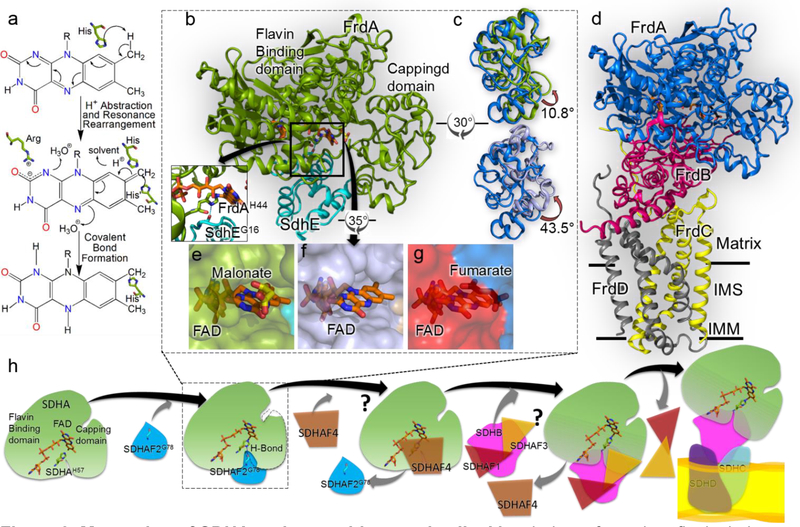
Figure 3. Maturation of SDHA and assembly complex II.
a Mecahnism of covalent flavinylation, as proposed by Walsh [49]. The first step is proton abstraction from the C8α of the isoalloxazine ring. The second step is resonance rearrangement to delocalize the resultant negative charge between N1 and C2. The third step is the attack by a histidyl ligand which results in formation of covalent bond. b The structure of the E. coli FrdA-SdhE assembly intermediate (PDB 6B58, [21••]). FrdA is colored green and SdhE is colored cyan. Inset The hydrogen bond between bacterial SdhEG16 of the RGxxE motif and FrdAH44 shows how SdhE orients the histidyl ligand. c Overlay of the capping domains of assembled FrdABCD complex (blue, PDB 3P4P, [50]) with the position observed in the FrdA-SdhE intermediate (top, green, PDB 6B58, [21••]) or SdhA-SdhE intermediate (bottom, purple, PDB 6C12 [42•]). A rotation of 10.8° and 43.5° is observed in the capping domains, respectively. d The structure of the E. coli fumarate reductase (PDB 3P4P [50]) with FrdA oriented as in Fig. 3b. IMM is inner mitochondrial membrane and IMS is inter-mitochondrial space. e, f Surface representation view of the FrdA-SdhE (PDB 6B58) and SdhA-SdhE (PDB 6C12) intermediates [21••,42•] highlights a tunnel from the solvent to the active site. g A similar surface representation of the FrdA subunit in the context of assembled complex II (PDB 3P4P, [50]) indicates that this tunnel closes following assembly. h A schematic representation of the proposed SDH assembly pathway. The details of SDHA-SDHAF2 interaction are known but structural states of interaction between assembly factors SDHAF1, SDHAF3 and SDHAF4 with SDH subunits is still unclear, and marked as “?”. Understanding how these assembly factors work is the focus of current research in the field.