Figure 1.
Characterization of the Novel Antisense lncRNA PTB-AS at the PTBP1 Locus
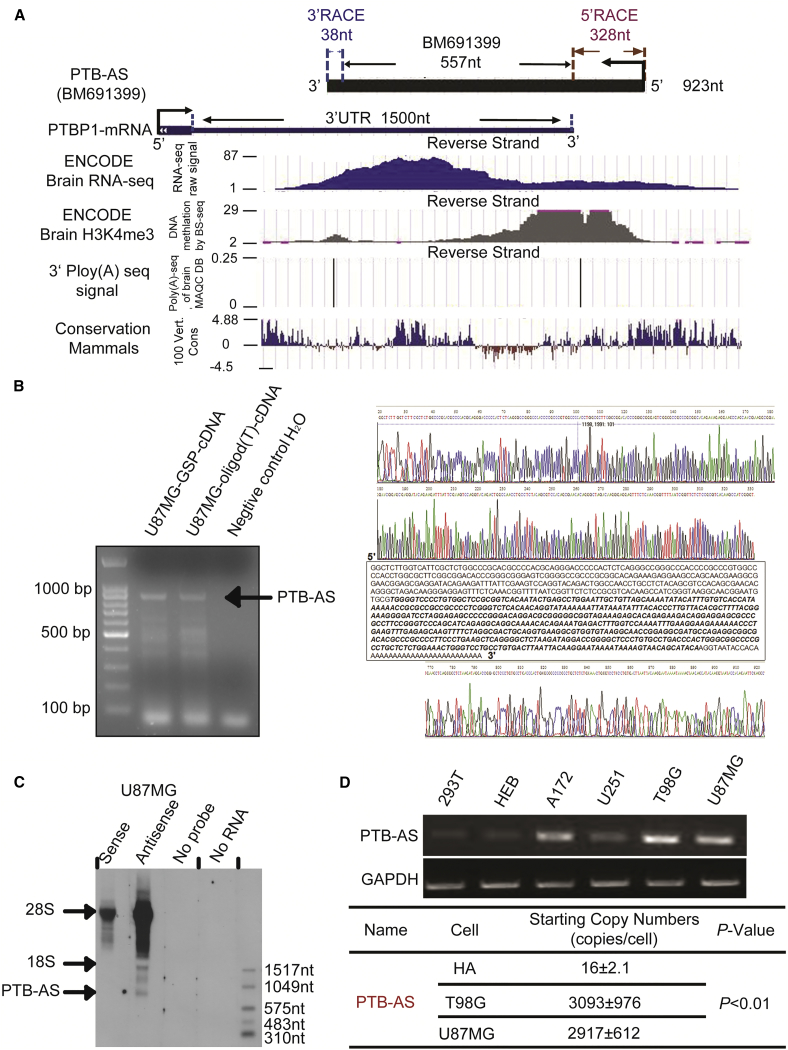
(A) Genome-wide discovery of PTB-AS. PTB-AS (923 nt), derived from the transcript BM691399 (557 nt), was amplified by RACE and overlapped with the PTBP1-3′ UTR. RNA sequencing (RNA-seq) can directly define the transcribed regions of the genome, and the raw signal here shows the primary structure of PTB-AS. Chromatin marks of transcription initiation (histone H3 lysine 4 trimethylation [H3K4me3]) defined the beginning of PTB-AS, and sequencing of polyadenylation ends (3′ Poly(A)-seq) defined the precise ends of these transcripts. The 100 Vert. Cons scores describe the conservation of PTB-AS in mammals. All of these raw data were analyzed using the UCSC genome browser. (B) Identification of the full length of PTB-AS by strand-specific RT-PCR. The total RNA of U87MG cells was extracted. The first lane indicates products that used the GSP for RT as the template. The second lane indicates products that used the oligo (dT) primer for RT as the template. The third lane indicates products that used H2O for RT as the template. The arrow shows the direction of the bands of PTB-AS. The raw sequencing spectrogram of RACE for PTB-AS is shown: the upper one is for 5′ RACE, and the lower one is for 3′ RACE. (C) Northern blot results indicate the full length of PTB-AS. An antisense probe hybridized with PTB-AS RNA, whereas the sense or no-probe and no-total RNA served as negative controls. The single band indicated by the arrow is PTB-AS. (D) Two normal human cell lines and four glioma cell lines were used to extract total RNA, which was reverse transcribed. Semiquantitative PCR was performed to identify the relative expression of the PTBP1 NAT. GAPDH served as the internal parameter reference. The copy number was detected and calculated to quantify the expression of PTB-AS in HA, T98G, or U87MG cell lines, using absolute qPCR.