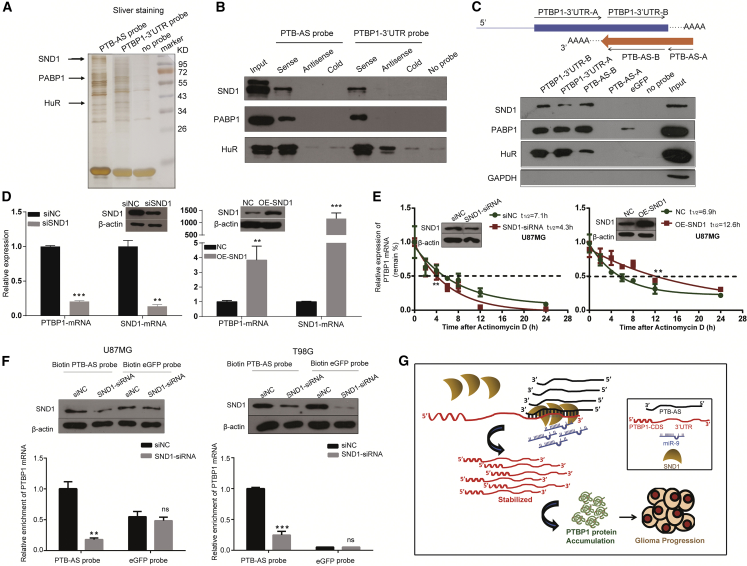
Figure 6.
SND1, Which Is Necessary for the Interaction of PTBP1 mRNA and PTB-AS, Promotes PTBP1-mRNA Stability
(A) The search for RNA-binding proteins using affinity purification. Biotin-labeled probes corresponding to full-length PTB-AS or the 3′ UTR of PTBP1 were incubated with T98G cell lysates and analyzed by SDS-PAGE, and the proteins were then silver stained. The negative control group used no probe. The pull-down samples were analyzed by mass spectrometry. (B) RNA pull-down assays verified the binding capacity of candidate proteins to PTB-AS and PTBP1-3′ UTR in T98G cells. (C) Identification of specific regions in PTB-AS and PTBP1-3′ UTR bound by candidate proteins. The schematic representation of different segments of transcripts used as probes in affinity purification reactions is shown above. The biotin-labeled EGFP probe as the negative control probe did not bind to any proteins. The probes were incubated with T98G cell lysates. Input protein came from the cell lysates before probes were incubated. GAPDH, as a negative control protein, did not bind to any probes. (D) Knocking down or overexpressing SND1 dramatically changed the expression of PTBP1 mRNA. U87MG cells were infected with siRNA (or siNC) or plasmid (or empty plasmid) targeting SND1, to manipulate the expression of SND1. Western blot analysis demonstrated the SND1 protein level. PTBP1 and SND1 transcripts were measured by quantitative real-time PCR, n = 3. *p < 0.05; **p < 0.01, ***p < 0.001 (Student’s t test). (E) U87MG cells expressing control siRNA or SND1 siRNA- or mock-vector (NC) or pcDNA4.0-SND1 plasmid (OE-SND1) were treated with actinomycin D (5 μg/mL) for the indicated periods of time. Total RNA was analyzed by RT-qPCR to examine the mRNA half-life of PTBP1. Data shown are the mean ± SD, n = 3. *p < 0.05, **p < 0.01 (two-tailed t test). The analyses used nonlinear regression (one-phase decay curve fit) to determine the half-life. (F) Biotin pull-down demonstrates that knockdown of SND1 weakens the binding between PTB-AS and PTBP1-3′ UTR. Cell lysates of U87MG cells and T98G cells expressing control siRNA or SND1-siRNA were incubated with in vitro synthesized biotin-labeled PTB-AS probe or EGFP probe for the biotin pull-down assays, followed by real-time qPCR analysis to examine PTBP1 mRNA levels. Total proteins from cell lysates were subjected to western blot with SND1 antibodies. Data shown are the mean ± SD (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001 (Student’s t test). (G) A schematic illustration of the proposed model depicting the role of PTB-AS in regulating PTBP1 mRNA stability. PTBP1 was significantly upregulated in glioma as a result of the dual-protection of PTB-AS; PTB-AS protected PTBP1 mRNA from inhibition by miR-9 through the masking effect; and SND1 dramatically stabilized PTBP1 mRNA through robustly enhancing the interaction of PTB-AS and PTBP1 mRNA.