Figure 3.
Spectrum of DLL1 Pathogenic Variants and Functional Interpretations
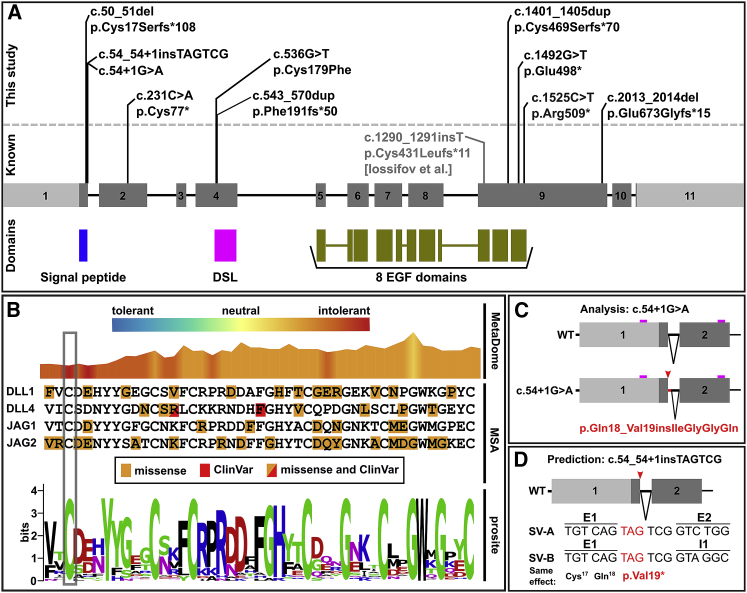
(A) Schematic of the DLL1 gene with DLL1 variants annotated. A previously described LoF variant is indicated in gray.23 The DLL1 protein domains are listed below.
(B) Sequence and variant characteristics of the DSL domain. The missense variant GenBank: NM_005618.3; p.Cys179Phe of I14 is highlighted in gray. Top: visualization of the MetaDome score.26 Center: multi sequence alignment (MSA) of the DSL domain of genes annotated with DSL domain by Prosite (DLL1, DLL4, JAG1, JAG4). Positions with missense variants in gnomAD version 2.1 as well as pathogenic ClinVar variants are labeled (no frameshift and nonsense variants are present in gnomAD). Bottom: Conservation of the DSL domain (Prosite entry PS51051). Cysteine is conserved among DSL domains. The MetaDome score indicates the deleterious nature of any amino acid changes at this position.
(C) Scheme of the consequence of the splice site variant identified in I12. Alternative splicing of the allele carrying c.54+1G>A leads to the retention of 12 intronic base pairs (bp), resulting in an in-frame insertion of four amino acids (p.Gln18_Val19insIleGlyGlyGln).
(D) Predicted consequence of the insertion c.54_54+1insTAGTCG identified in I13. Human Splicing Finder29 predicted that the insertion c.54_54+1insTAGTCG leads to an alternative donor splice site after the insertion, resulting in two different predicted transcript forms. Both lead to a premature stop codon. Boxes present exons, lines present introns; light gray boxes: untranslated regions; dark gray boxes: coding sequence; small boxes in magenta in (C): location of the sequencing primers; red arrows: location of the splice variants; black triangles: intronic sequence removed by splicing; SV-A: splice variant A; SV-B: splice variant B. E1 and E2 represent sequence from exon 1 and 2 respectively, I1 represents intron 1.