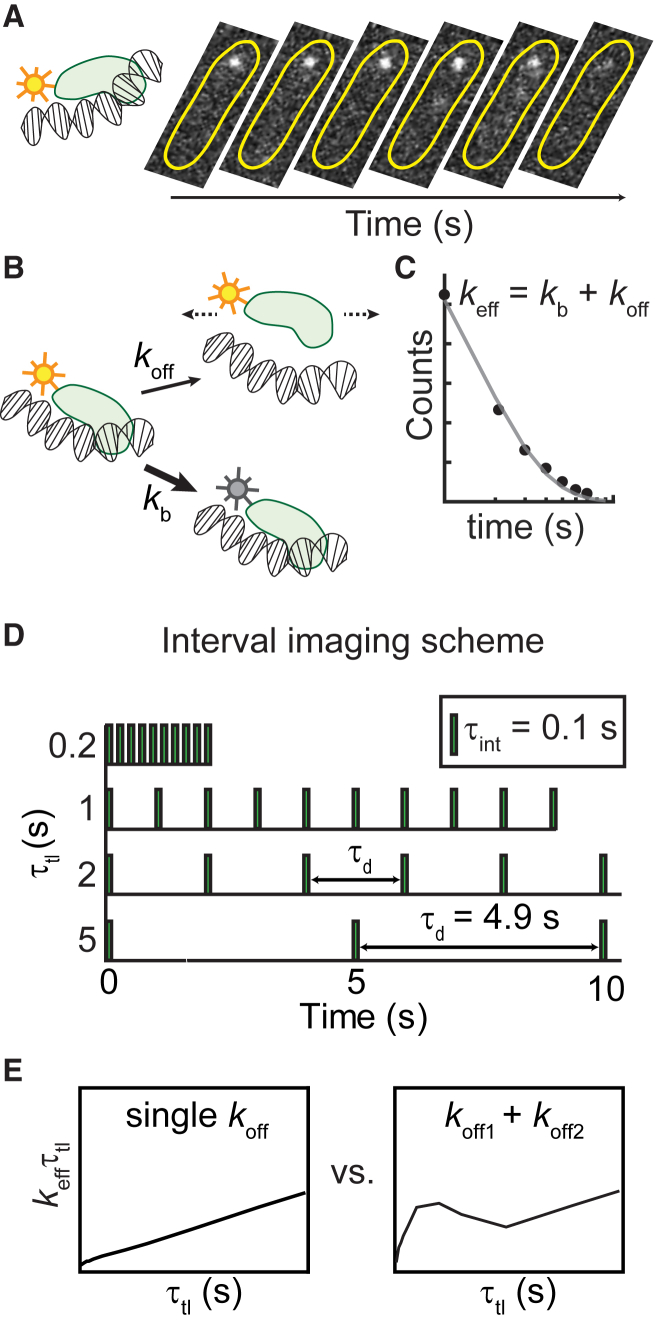
Figure 1.
Experimental approach for characterizing kinetic heterogeneity of protein binding in live cells using single-molecule fluorescence imaging. (A) The protein of interest is tagged with a fluorescent protein. When the protein binds to the DNA substrate, its fluorescence signal appears as a diffraction-limited focus that can be tracked in real time. Subsequent dissociation results in the disappearance of the focus and a redistribution of the fluorescence signal throughout the cell. Yellow outlines illustrate the bacterial cell membrane. (B) The loss of fluorescence is attributable to either dissociation or photobleaching of the chromophore. (C) Cumulative residence time distribution (CRTD) is constructed from binding durations of thousands of molecules. Fitting the exponential function (Eq. 1) to CRTD yields an effective rate keff, which is the sum of off rate (koff) of the protein of interest and photobleaching rate (kb) of the fluorescent probe (23). (D) To deconvolute kb and koff, excitation and integration durations (τint) can be spaced with various dark intervals (τd). (E) Through exponential analyses, CRTDs obtained at various intervals result in keffτtl plots, which are indicators of kinetic heterogeneity (23). A single kinetic population yields a straight line, whereas deviations from linear fits indicate the presence of a second kinetic subpopulation. For a single kinetic population, the slope is the off rate, and the y intercept is proportional to the photobleaching rate. To see this figure in color, go online.