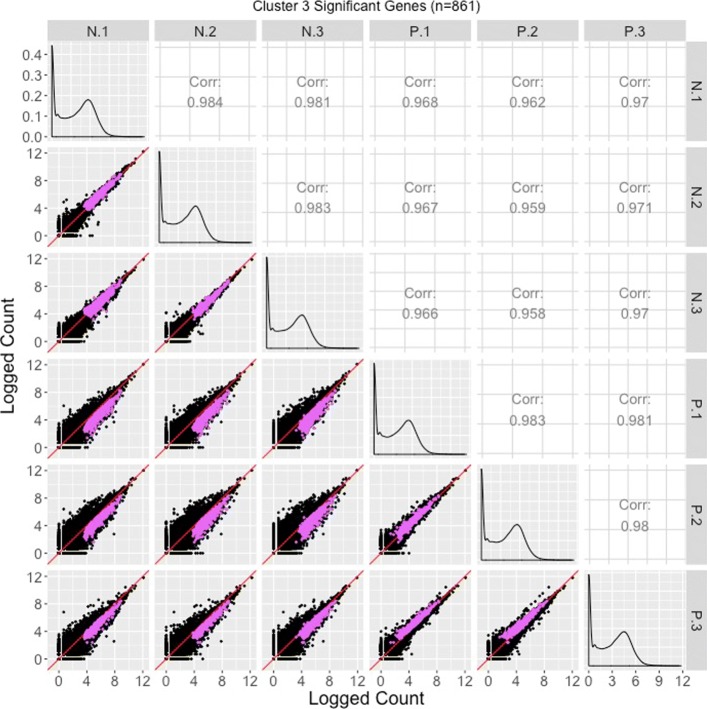
Fig. 13.
Cluster 3 significant genes from the soybean iron metabolism data overlaid on a scatterplot matrix. Example of using a scatterplot matrix to assess DEG calls from a model in the iron-metabolism soybean dataset. There were 861 significant genes in Cluster 3 after performing a hierarchical clustering analysis with a cluster size of four (Fig. 2). These significant genes are overlaid in pink on the scatterplot matrix. For the most part, they follow the expected patterns of differential expression with pink points falling along the x=y line in the scatterplots between replicates, but deviating from the x=y line in the scatterplots between treatments. The deviation consistently demonstrates higher expression in the N group than in the P group. The scatterplot between replicates P.1 and P.3 shows slightly higher expression in P.3, and the scatterplot between replicates P.2 and P.3 also shows slightly higher expression in P.3. Hence, these pink points seem to represent genes that were significantly overexpressed in the N group, but with slight inconstencies in the replicates in the P group, which matches what we saw in the parallel coordinate plots in Fig. 2. One difficulty with plotting such a large number of DEGs onto the scatterplot matrix is that overplotting can obscure our inability to determine how many DEGs are in a given location. This is why we might also view these genes individually in litre plots (Fig. 10e and f)