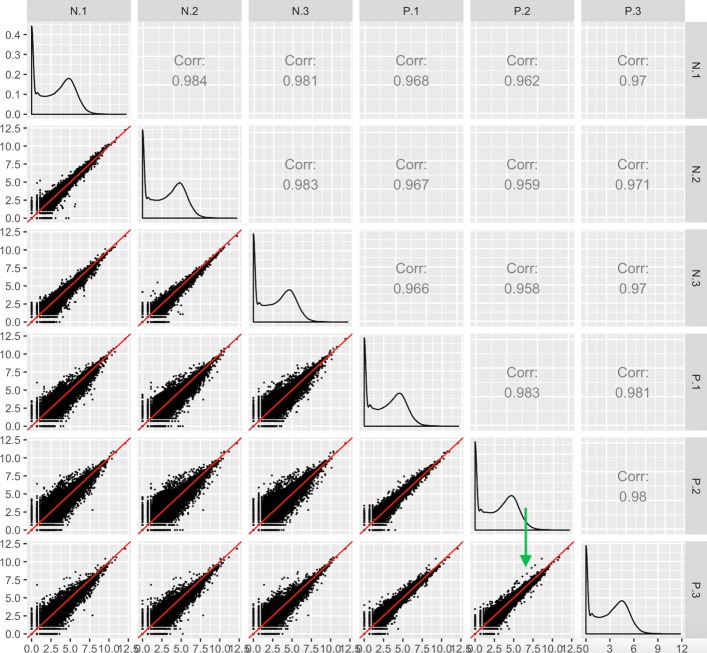
Fig. 7.
Finding unexpected patterns in RNA-seq data using scatterplot matrices. Scatterplot matrix of RNA-seq read counts from soybean leaves after exposure to iron-sufficient (treatment group P) and iron-deficient (treatment group N) hydroponic conditions [24]. We observe the expected structure of treatment pairs showing larger variability around the x=y line than replicate pairs. However, we notice a pronounced streak structure in the bottom-right scatterplot (green arrow) that compares two replicate samples from the iron-sufficient group. The genes in the streak structure have large read counts that deviate in a parallel fashion from the x=y line