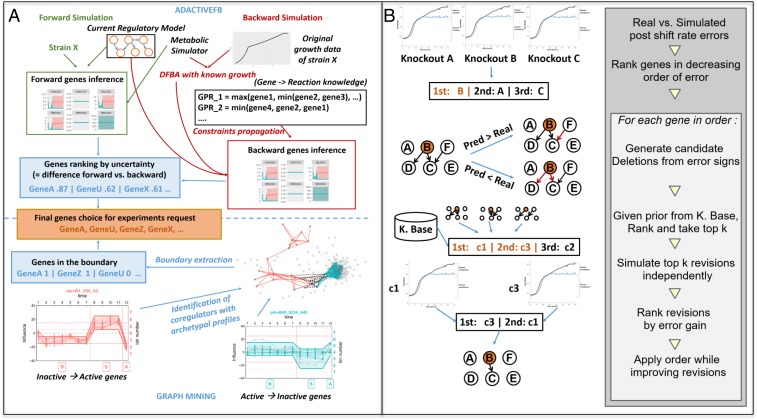
Fig. 4.
(A) The active learning tool AdactiveFB for selecting experiments identifies the most uncertain genes in a regulatory model using forward-backward simulation. It infers phenotype-driven distributions about genes in the regulatory model, which are then compared to gene distributions obtained from simulation. AdactiveFB first applies DFBA with fixed growth rates (instead of growth maximization) to estimate metabolic genes activity from growth curves by finding the metabolic reaction bounds associated to observed growth rates. It then propagates these constraints to the metabolic gene distributions in the regulatory model, before finally propagating them to regulatory genes using Bayesian inference. (B, Left) The tool CoRegMine used the Brauer dataset to form a graph of gene–target relationships. The graph mining tool MinerLC* selects genes belonging to a dense subgraph of the coregulation graph that have antagonist influence profiles along this time series, i.e., inactive → active vs. active → inactive denoted, respectively, by the red and blue nodes and links in the figure. Regulators at the border of those two subgraphs (i.e., nodes with active → inactive profiles which are neighbors—denoted by black links—of nodes with inactive → active profiles in the coregulation graph) are selected. (B, Right) The tool Adarev for model refinement. From the set of growth curves derived from for a set of knocked out genes, a set of prediction vs. observed postshift growth rate errors are computed and used to rank genes for removal. This ranking is used greedily to apply revisions starting with the most promising ones, iteratively validating the proposed changes as long as new predictions in the updated models are better than previous ones, in terms of postshift growth rate error reduction gain (S11).