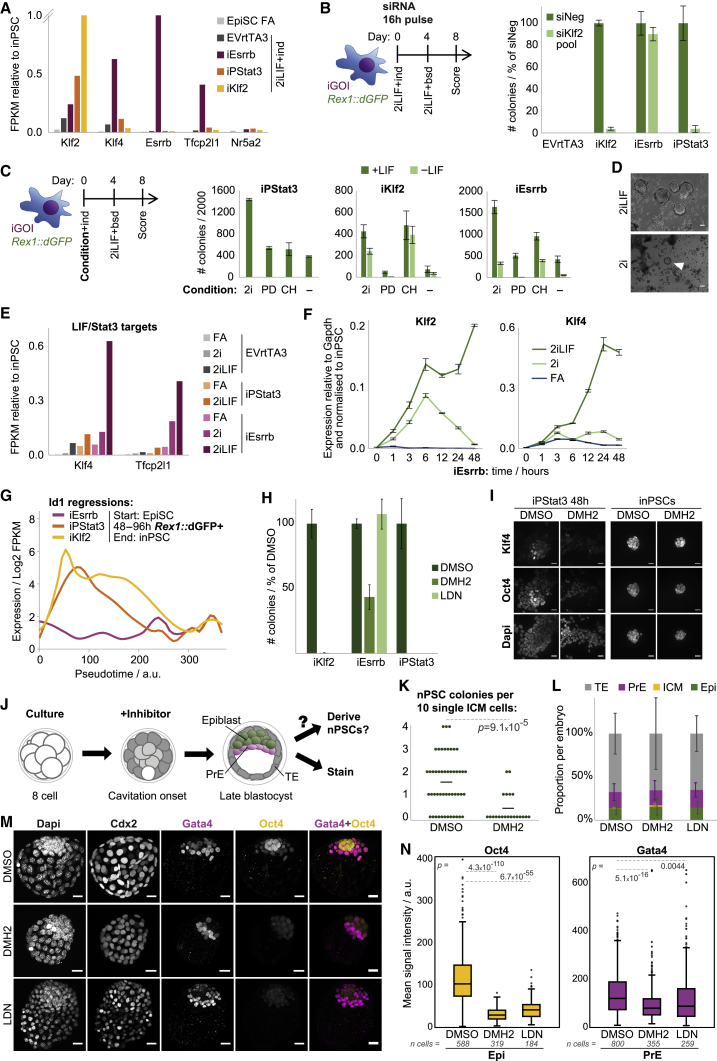
Figure 5.
Routes Have Distinct Genetic and Signal Requirements
(A) Gene expression after 24 h relative to inPSCs. y-axis: iEsrrb, Esrrb = 3.32; iKlf2, Klf2 = 8.30.
(B) KD was performed at reprogramming onset with a single pulse of small interfering (si) RNA. Mean inPSC colonies scored on day 8 are presented ± SD (n = 3).
(C) Reprogramming was induced under different conditions from days 0–4 and then selected in 2iLIF+blasticidin. inPSC colonies scored on day 8 are presented as mean ± SD (n = 3). 2i, PD+CH.
(D) Phase images of iEsrrb on day 8 in 2iLIF+blasticidin after reprogramming from days 0–4 in 2iLIF+dox or 2i+dox as indicated. The arrowhead indicates an inPSC colony.
(E) Expression of LIF/Stat3 target genes 24 h after driver induction under the indicated conditions.
(F) Timecourse RT-qPCR analyses of iEsrrb EpiSCs under the indicated conditions + dox. Mean expression is displayed ± SD (n = 3).
(G) LOESS regression fit lines summarize Id1 kinetics during reprogramming, computed from log2 FPKM versus pseudotime for single cells (Figure S2C).
(H) 3 μM DMH2, 0.6 μM LDN, or DMSO were applied to reprogramming in 2iLIF+dox/GCSF from days 0–4, and then inPSCs were selected in 2iLIF+blasticidin. inPSC colonies scored on day 8 are presented as mean ± SD (n = 3).
(I) Immunofluorescent staining after 48 h of inhibitor treatment for iPStat3 reprogramming in 2iLIF+GCSF or for previously established iPStat3 inPSCs in 2iLIF. Scale bars, 20 μm.
(J) Schematic summarizing BMP inhibitor treatment of pre-implantation embryos.
(K) Quantitative nPSC derivation following embryo treatment with DMSO or 3 μM DMH2. nPSC colonies were scored per 10 single ICM cells. Black line, mean. DMSO, n = 7; DMH2, n = 8 embryos.
(L–N) Late blastocysts were stained for Cdx2, Gata4, and Oct4 following treatment with DMSO, 3 μM DMH2, or 0.3 μM LDN.
(L) Mean cell number per lineage ± SD, presented as a proportion of the total cells per embryo. DMSO, n = 23; DMH2, n = 18; LDN, n = 7 embryos.
(M) Representative maximum intensity Z-projections and indicated merge. Scale bars, 20 μm.
(N) Quantification of immunofluorescent signal for Oct4 in Epi nuclei and Gata4 in PrE nuclei, presented as box-and-whisker plots. DMSO, n = 23; DMH2, n = 18; LDN, n = 7 embryos.
See also Figure S5.