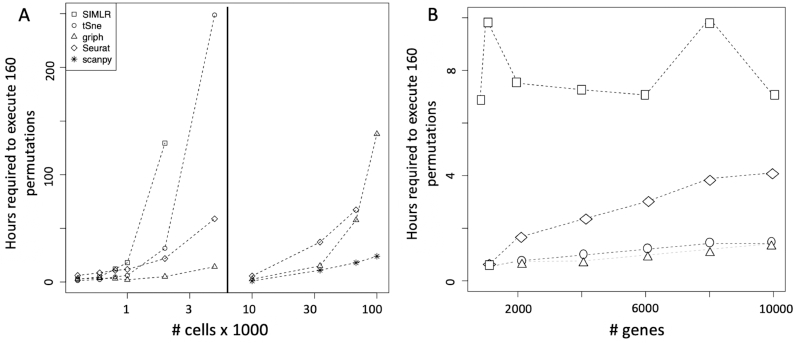
Figure 7:
Scalability analysis of the clustering tools implemented in rCASC. A, Time required to perform 160 permutations as function of increasing number of cells on ∼20,000 genes. Left: SIMLR, t-SNE, Seurat, and griph clustering up to 5,000 cells was executed on a SeqBox [9] (1 x CPU i7–6770HQ 3.5 GHz [8 cores], 32 GB RAM, 1 TB SSD). Right: Seurat, griph, and scanpy analyses were extended until 101,000 cells using an SGI server (10 x CPU E5–4650 2.4 GHz [16 cores], 1 TB RAM, 30 TB SATA raid disk). B, Time required to perform 160 permutations as function of increasing number of genes on a set of 800 cells, analysis performed on a SeqBox.