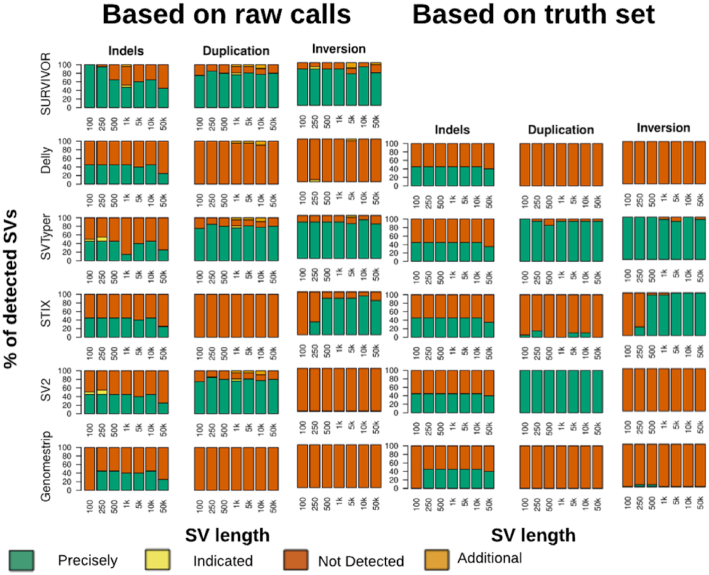
Figure 1:
Evaluation of Illumina-like reads to assess the SV genotyper ability to re-identify insertions, deletions, duplications, and inversions over different size ranges (x-axis). The colors indicate the SVs being detected/genotyped by the respective SV genotypers. They were classified as either precisely, indicated, not detected, or falsely identified (see Methods). For the SVs genotyped on the basis of SV calls (left) we used SURVIVOR, which is a union set of Delly, Lumpy, and Manta, to generate the VCF file as an input for the SV genotypers. Noteworthy, Delly and SVtyper can genotype more SVs, given the custom information from their respective callers, Delly and SVTyper, respectively. When the truth SV set is provided as a start point (right panel) we see marginal improvements across the SV genotyping methods while maintaining the overall trend.