Figure 2.
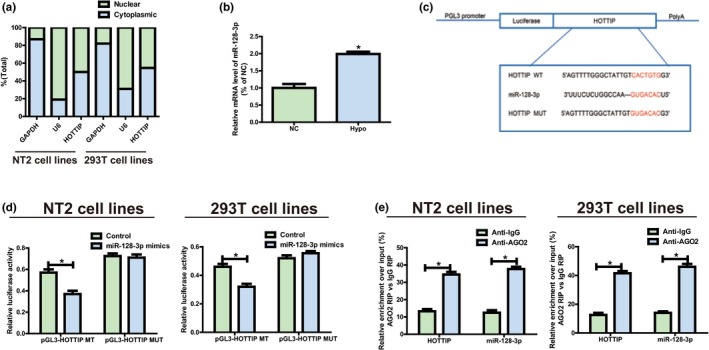
HOTTIP directly interacts with miR‐128‐3p. (a) Distribution characteristics of HOTTIP in NT2 and 293T detected via qRT‐PCR. (b) MiR‐128‐3p expression level is decreased in Hypo patients compared to the normal controls. (c) Bioinformatics evidence that miR‐128‐3p binds to HOTTIP’s 3’‐UTR. The speculated miRNAs recognition sites that are cloned into the luciferase gene downstream are named as pGL3‐HOTTIP‐WT. Bottom: Mutant HOTTIP sequences that produce the mutant luciferase reporter constructs gain the name of pGL3‐HOTTIP‐MUT. (d) Detection of luciferase activity in NT2 and 293T following co‐transfection with plasmids (pGL3‐HOTTIP‐WT or pGL3‐HOTTIP‐MUT) and miRNA mimics via dual‐luciferase reporter assay. (e) RIP assays are performed in NT2 and 293T to figure out whether HOTTIP is present in ribonucleoprotein complexes with miRNAs. QRT‐PCR is adopted to measure miR‐128‐3p and HOTTIP expression levels. Data are presented as mean ± SD. *p ≤ .05, Student’s t‐test
