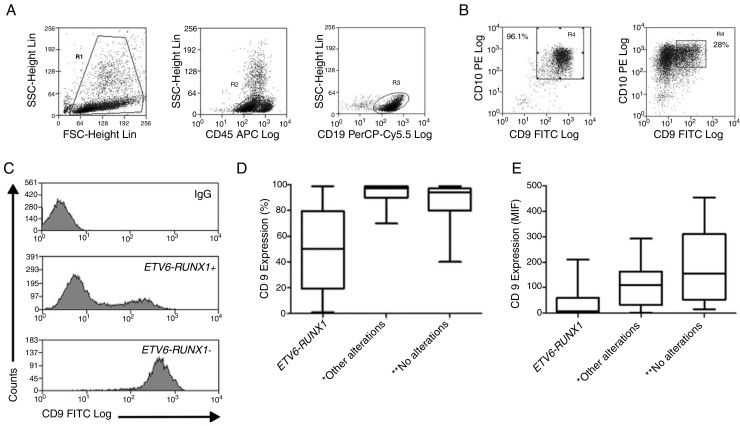
Figure 2.
CD9 expression in blast cells analyzed by FC and correlation between the CD9 percentage/MFI-r and the major molecular alterations in BCP-ALL. (A) Upper dot plots show the analysis strategy used to evaluate CD9 expression in the blast population, which was defined by the R1 (viable cells) + R2 (CD45 negative/low or intermediate) and R3 regions (CD19 positive cells). (B) R4 regions represent the percentage of CD9 positive blast cells. The left dot plot represents patients with a CD9 high-percentage expression (96%) and the right dot plot represents patients with a CD9 low-percentage expression (28%). (C) CD9 expression evaluated by ratio median fluorescence intensity (MFI-r) in blast population. In the upper histogram, non-specific IgG control histogram. The middle panel represents the low CD9 MFI-r expression in a patient ETV6-RUNX1+. The last panel represents the high CD9 MFI-r expression in a patient ETV6-RUNX1−. (D) CD9 percentage according to the common alterations (high hyperdiploidy, TCF3-PBX1, BCR-ABL1, KMT2A rearrangements) in BCP-ALL. (E) CD9 MFI-r, according to the common alterations in BCP-ALL. Horizontal lines inside the bars represent the median CD9 expression percentage or MFI-r obtained from the studied cases. *Other alterations refer to MLL-r (n = 16); TCF3-PBX1 (n = 4); BCR-ABL1 (n = 1). **No alterations were identified with the screening methods used in this study.