Fig. 6.
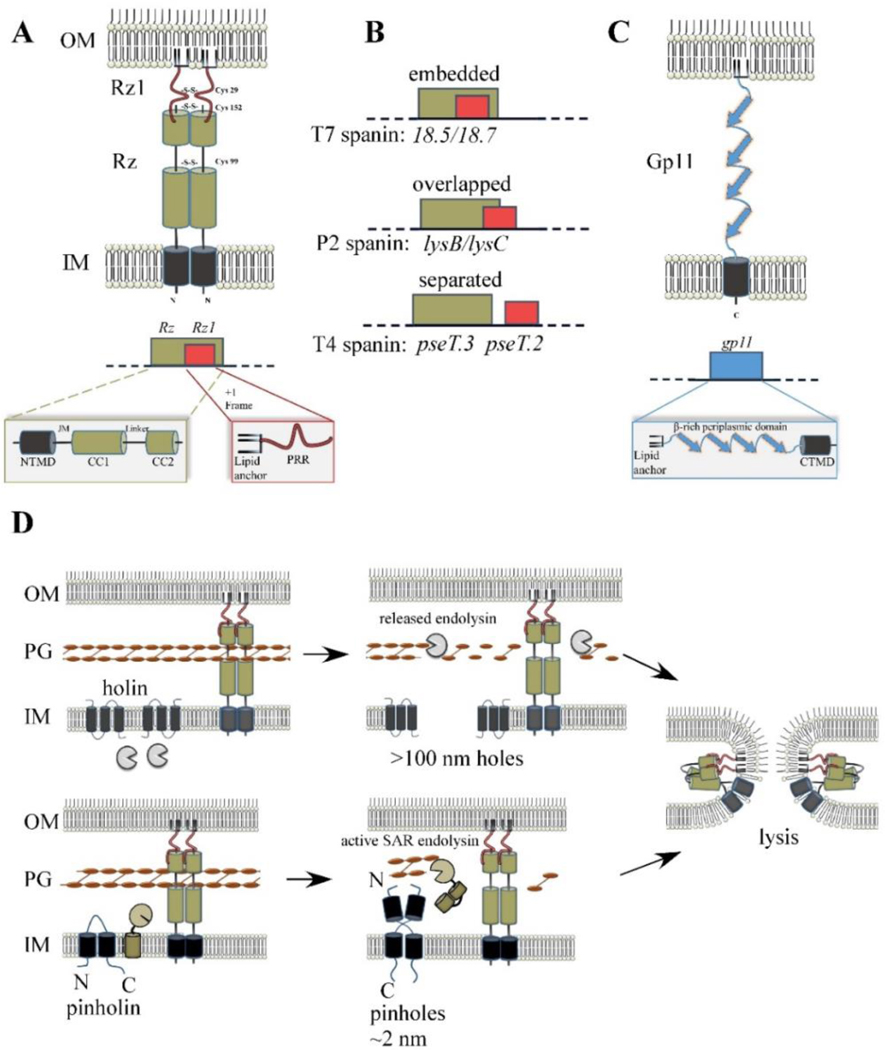
Spanin products, genes, and model for function. (A) Cartoon of λ spanin complex, comprised of Rz and Rz1, is shown within the cell envelope. The homotypic intermolecular disulfide linkages are denoted. Below: The Rz1gene is embedded within the +1 reading frame of Rz. Functional domains of the spanin complex that has been identified by genetic analysis are denoted in at relative positions respective to the predicted structural features. NTMD = N-terminal transmembrane domain. JM = juxtamembrane region, CC1 = alpha helix with predicted coiled-coil domains, CC2 = distal coiled-coil domain, PRR = proline-rich region. (B) Examples of different spanin genetic architectures, classified on the degree of overlap of the o-spanin gene (red) within the i-spanin (green). (C) The u-spanin gene product, exemplified by gp11 of T1. Below: Predicted domains of the gp11 product. (D) Model for spanin function following the canonical holin-endolysin (top) or pinholin-SAR endolysin pathway (bottom). For both pathways, spanins activate after PG degradation and undergo a conformational change that causes fusion of the IM and OM. This removes the topological barrier of the OM at the last step of lysis, which results in the release of phage progeny.
