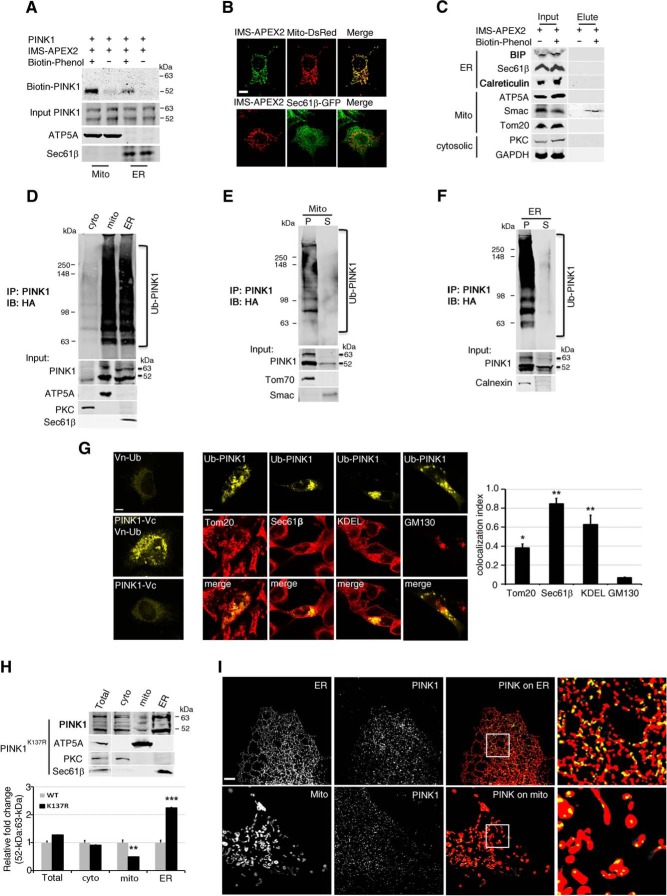
Figure 4.
Ub-PINK1 is found in both the ER and mitochondria. A, HeLa cells were cotransfected with PINK1 and IMS-APEX2, purified mitochondrial (mito) and ER fractions were used in the sequential pull-down. This experiment was repeated once independently with similar results. B, Mito-DsRed and Sec61β were used as mitochondrial and ER outer membrane markers, respectively. IMS-APEX2 specifically colocalized with mitochondrial marker Mito-DsRed in COS-7 cells. Scale bar, 10 μm. C, 293T cells were cotransfected with PINK1 and IMS-APEX2, labeled with biotin-phenol and processed for IP. Biotin-labeled protein was detected in elutes that was pulled-down by streptavidin magnetic beads using the total cell extract (Input). BIP, Sec61β, and calreticulin were used as ER markers; ATP5A, Smac, and Tom20 were used as mitochondrial markers; PKC and GAPDH were used as cytosolic markers. This experiment was repeated once independently with similar results. D, Ub-PINK1 IP followed by IB analysis of purified subcellular fractionation isolated from cotransfected PINK1 and HA-Ub HeLa cell exposed to MG132 (n = 3). E, F, Alkaline extraction (0.1 m Na2CO3, pH 11) of purified mitochondrial (E) or ER fractions (F) from the same cells as in Figure 3D followed by IP and IB analysis (n = 3). P, Particulate; S, supernatant. Tom70, Smac, and calnexin are markers of mitochondrial outer membrane, mitochondrial interspace and ER membrane integral proteins, respectively. G, Left, The Venus signal (yellow) was only observed in HeLa cells cotransfected with Vn-Ub and PINK1-Vc constructs. Middle, Ub-PINK1 signal (yellow) in HeLa cells coexpressing Vn-Ub and PINK1-Vc and immunostained with antibody against Tom20, Sec61β, KDEL, or GM130 (red). Scale bar, 10 μm. Right, Quantification of Ub-PINK1 and organelle marker colocalization index. *p < 0.005, **p < 0.001 difference from GM130 (one-way ANOVA, F(3,8) = 22.83, p < 0.001, followed by a Holm-Sidak's post hoc test). H, Top, PINK1 IB of purified subcellular fractions of HeLa cells expressing PINK1K137R. Bottom, PINK1 52 kDa:63 kDa ratio relative to WT PINK1 expressing cells. **p < 0.001, ***p < 0.0001 difference from WT controls (two-way ANOVA, interaction mutation × subcellular fraction: F(3,16) = 58.35, p < 0.001, followed by a Holm–Sidak's post hoc test). I, SIM imaging analysis was performed similar to that in Figure 3B, except that cells were transfected with PINK1K137R. Scale bar, 100 μm.