Fig. 7. Simulation analyses of the docking of PAPS and pregnenolone into the active site of SULT2B1b allozymes.
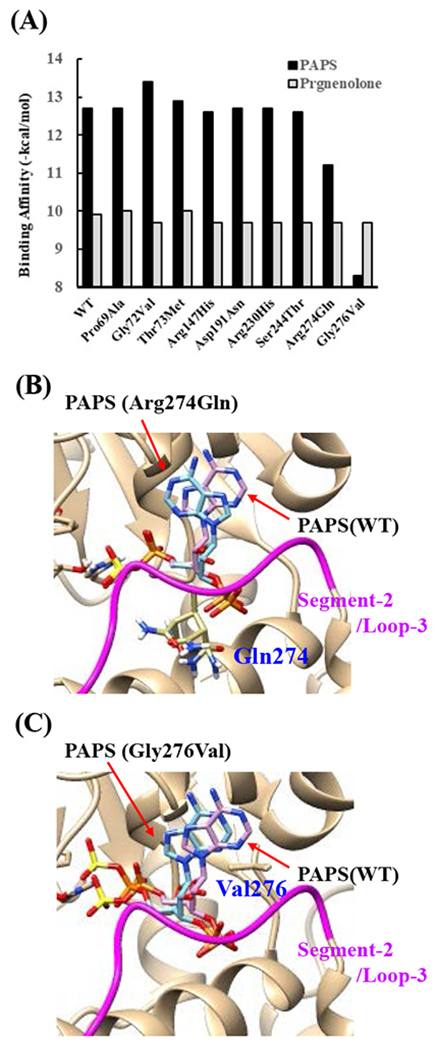
(A) Minimum binding energy of PAPS (black scale) and pregnenolone (grey scale) in the active site of SULT2B1 allozymes. Conformation of mutated amino acid residues of allozymes simulated using the Dunbrack backbone-dependent rotamer library was used for docking analyses as the template protein structures. (B) Stereo view of the PAPS docked into the active site of Arg274Gln. Superpositions of PAPS docked in Arg274Gln and wild-type are shown in blue and purple backbone, respectively. (C) Stereo view of the PAPS docked into the active site of Gly276Val. Superpositions of PAPS docked in Gly276Val and wild-type are shown in blue and purple backbone, respectively.
