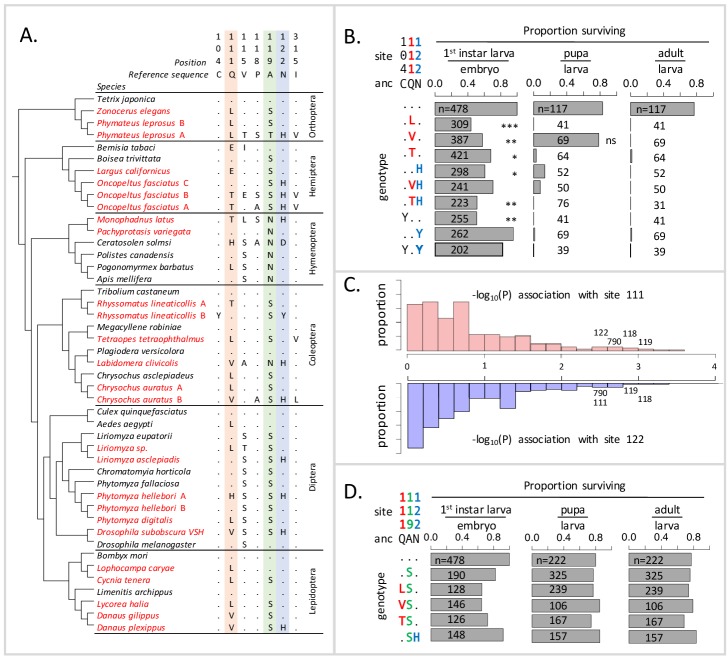
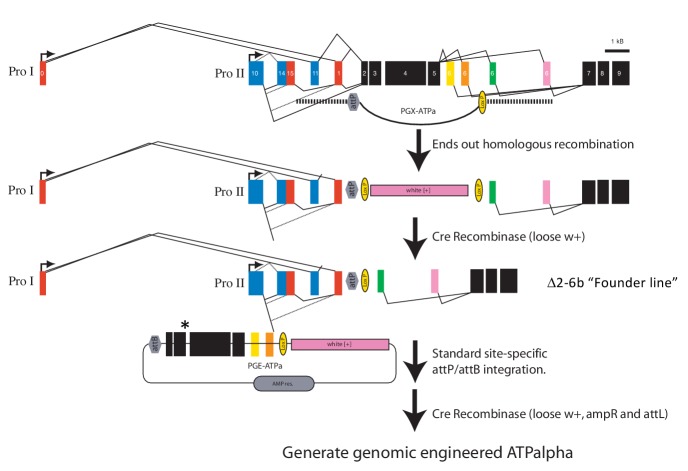
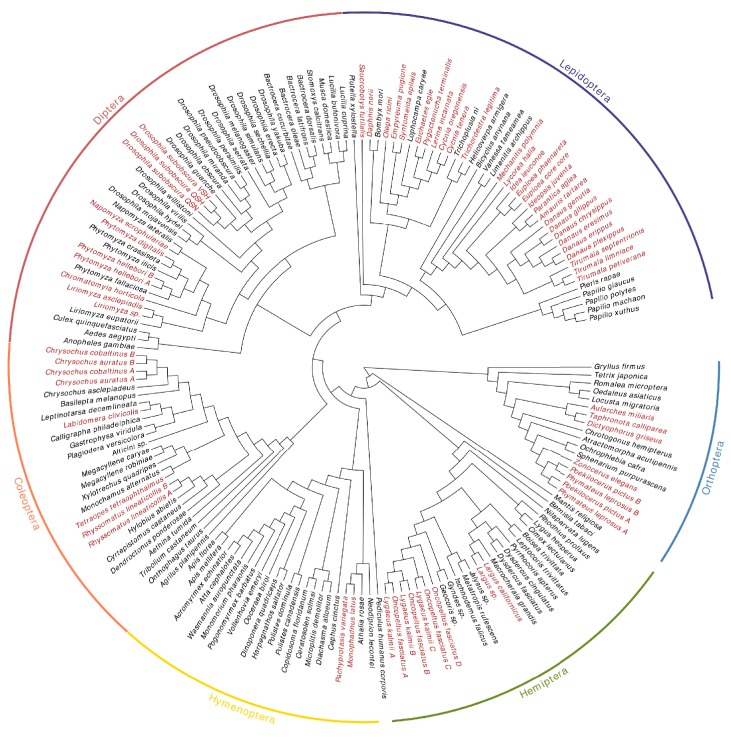
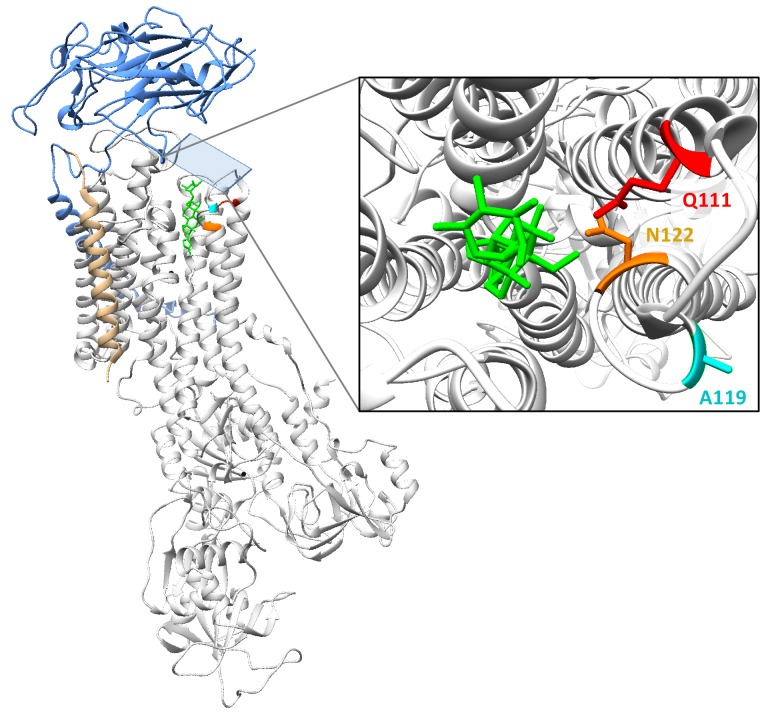
Figure 1. Deleterious effects of substitutions at positions 111 and 122 are ameliorated by the permissive substitution A119S.
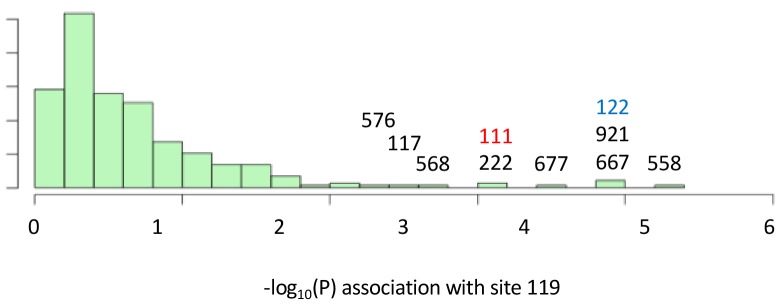
(A) Patterns of amino acid substitution at sites implicated in CG sensitivity near the H1-H2 transmembrane domain of ATPα1 in representative species from six insect orders (see Supplementary file 1 for all species). CG-adapted species names are shown in red. Dots indicate identity with the ancestral reference sequence and letters indicate derived amino acid substitutions. Positions 111 and 122, highlighted in pink and blue respectively, are hotspots of frequent parallel substitution. C104Y and N122Y represent rare substitutions associated with ATPα1 duplication (Zhen et al., 2012). Position 119 (highlighted in green) had not been previously implicated in CG sensitivity, but is identified as a candidate permissive substitution in subsequent analyses. (B) Viability of homozygous first instar larvae, pupae and adults for the first series of engineered substitution lines. Dots indicate identity with the ancestral reference at sites C104, Q111, and N122. Plotted is the proportion of surviving homozygous mutant offspring (i.e. EYFP-), scaled by the proportion for the wild-type control. The number of individuals assayed is indicated in each case. Survival odds relative to the wild type-strain (…) was tested using a Fisher’s Exact Test, with adjusted P-values indicated as: ***p<0.001; **p<0.01, *p<0.05. All larvae to pupa survivorship are significantly lower than wild-type (p<1e-5) except for Q111V. All larvae to adult survivorship are significantly lower than wild-type (p<1e-5). (C) Distributions of P-values for the strength of the phylogenetic correlation between sites 111 (above) or 122 (below) and 270 non-singleton amino acid variants in a multi-sequence alignment including 174 ATPα1 sequences representing 161 species. Sites 111 and 122 exhibit highly correlated evolution. Three additional sites in the lowest 5% of P-values shared in common between sites 111 and 122, including site 119, are labeled. (D) Viability of homozygous first instar larvae, pupae and adults for the second series of engineered substitution lines that include A119S relative to the wild-type control. Dots indicate identity with the ancestral reference at sites Q111, A119, and N122. The number of individuals assayed is indicated in each case. Despite slight reductions in viability, all reductions are not significant relative to wild-type line after multiple test correction.