Abstract
Prevention of Epstein-Barr virus (EBV) infection has focused on generating neutralizing antibodies (nAbs) targeting the major envelope glycoprotein gp350/220 (gp350). In this study, we generated 23 hybridomas producing gp350-specific antibodies. We compared the candidate gp350-specific antibodies to the well-characterized nAb 72A1 by: (1) testing their ability to detect gp350 using enzyme-linked immunosorbent assay, flow cytometry, and immunoblot; (2) sequencing their heavy and light chain complementarity-determining regions (CDRs); (3) measuring the ability of each monoclonal antibody (mAb) to neutralize EBV infection in vitro; and (4) mapping the gp350 amino acids bound by the mAbs using competitive cell and linear peptide binding assays. We performed sequence analysis to identify 15 mAbs with CDR regions unique from those of murine 72A1 (m72A1). We observed antigen binding competition between biotinylated m72A1, serially diluted unlabeled gp350 nAbs (HB1, HB5, HB11, HB20), and our recently humanized 72A1, but not gp350 non-nAb (HB17) or anti-KSHV gH/gL antibody.
Keywords: Epstein-Barr virus, gp350, neutralizing antibodies, complementarity-determining region, immunodominant, epitope, immunosuppression, infection
Introduction
Epstein-Barr virus (EBV) predominantly infects epithelial cells and B cells, reflecting the viral tropism and cellular ontogeny characteristic of most EBV-associated malignancies (Rickinson and Kieff, 2007b). Despite the fact that EBV infection is associated with more than 200,000 cases of a variety of human malignancies every year, and has significant public health impacts, there is no licensed vaccine to date (Cohen et al., 2011). The EBV glycoprotein gp350/220 (gp350) is a known target for a host’s virus neutralizing antibody (nAb) response upon natural EBV infection (Sashihara et al., 2009; Thorley-Lawson and Poodry, 1982; Weiss et al., 2017) or immunization, and thus has been tested as a viable target for vaccines and therapeutics in five clinical trials to prevent B cell infection (Gu et al., 1995; Haque et al., 2006; Moutschen et al., 2007; Rees et al., 2009; Sokal et al., 2007). However, not all of the potential nAb epitopes on gp350 have been identified or fully characterized.
EBV infects at least 90% of the human population globally, irrespective of geographical location. Currently, there are two models describing how initial EBV infection of human host cells occurs in vivo (Cohen, 2000). In the first infection model, the incoming virus first targets epithelial cells and engages with host ephrin receptor tyrosine kinase A2 via heterodimeric glycoproteins gH/gL (Chen et al., 2018; Zhang et al., 2018) or with host integrins via BMRF-2 (Chesnokova et al., 2009; Tugizov et al., 2003). This triggers fusion of EBV glycoprotein gB with the host epithelial cell membrane to enhance viral entry into the cytoplasm. This interaction is thought to occur in the oral mucosa; there, EBV undergoes lytic replication in epithelial cells to release virions that subsequently infect resting B cells in tonsillar crypts or circulating naïve B cells. In the alternative infection model, the incoming virus binds to B cells in the oral mucosa via host CD35 (Ogembo et al., 2013) and/or CD21 through its major immunodominant glycoprotein, gp350 (Fingeroth et al., 1984; Nemerow et al., 1989). The interaction between gp350 and CD35 and/or CD21 triggers viral adsorption, capping, and endocytosis into B cells (Tanner et al., 1987). This subsequently leads to the heterotrimeric EBV glycoprotein complex gp42/gH/gL binding to host HLA class II molecules to activate gB membrane fusion and infection of B cells (Connolly et al., 2011). Once infected, B cells typically remain latent and harbor the virus for life, but may also traffic back to the oropharynx, where EBV is amplified by lytic replication in epithelial cells, and shed into the saliva (Cohen, 2000). Thus, B cells are the main reservoirs for EBV reactivation and for the development of virus-related malignancies (Babcock et al., 1998). Novel strategies that could block interactions between EBV glycoproteins and cellular receptors that mediate viral infection could be beneficial in the development of effective antiviral therapies.
Antibodies are the first line of defense against viral infection and nearly all EBV-infected individuals develop nAbs directed to the ectodomain of EBV gp350 (Sashihara et al., 2009; Thorley-Lawson and Poodry, 1982; Weiss et al., 2017). A recent study showed that polyclonal serum antibodies against gp350 from naturally infected individuals or immunized animals block EBV infection of B cells in vitro better than antibodies against EBV gH/gL or gp42 (Bu et al., 2019). Thus, gp350 is a promising candidate for development of EBV vaccines against B cell infection; however, to make effective vaccines, the nAbs epitopes on the gp350 ectodomain must be identified and fully characterized. EBV gp350, a type 1 membrane protein, is composed of 907 amino acid (aa) residues. A single splice of the primary gp350 transcript deletes 197 codons between codons 501 and 699, and joins two fragments in frame, to generate the gp220 transcript. Both gp350 and gp220 are composed of the same 18-aa residue at the C terminus that is located within the viral membrane, a 25-aa residue at the transmembrane-spanning domain, and a large highly glycosylated N-terminal ectodomain, aa 1–841 (Tanner et al., 1988). The first 470 aa of gp350 are thought to be sufficient for binding CD21 in B cells, as demonstrated by a truncated gp350 (aa 1–470) blocking the binding of EBV to B cells and reducing viral infectivity (Tanner et al., 1988). The gp350-binding domain on CD21 maps to N-terminal short consensus repeats (SCRs) 1 and 2, which also bind to a bioactive fragment of complement protein 3 (C3d) (Nemerow et al., 1987a). A soluble truncated gp350 fragment (aa 1–470) and soluble CD21 SCR1 and SCR2 can block EBV infection and immortalization of primary B cells (Tanner et al., 1988). However, gp350 binding to CD35 is not restricted to N-terminal SCRs; it binds long homologous repeat regions as well as SCRs 29–30 (Ogembo et al., 2013).
There are at least seven unique CD21 binding epitopes located in the ectodomain of gp350 (Urquiza et al., 2005); at least one of these epitopes (aa 142–161) is capable of eliciting nAbs (Tanner et al., 1988; Urquiza et al., 2005). The aa residues 142–161 are also one of the binding epitopes for nAb 72A1 (Hoffman et al., 1980; Szakonyi et al., 2006). Using gp350 synthetic peptides binding to CD21 on the surface of a B-cell line, an additional gp350 epitope was identified in the C-terminal region of gp350 (aa 822–841), suggesting this region also involved in EBV infection of B cells (Urquiza et al., 2005). However, the role of other epitopes in eliciting nAbs has not been fully investigated to fully justify the recent use of partial sequence of gp350 ectodomain in the development of effective prophylactic vaccine (Cui et al., 2013; Kanekiyo et al., 2015). Furthermore, the exact aa residues that comprise the core binding epitopes capable of eliciting nAbs and non-nAbs have not been determined. Mapping the gp350 protein residues that define immunodominant epitopes, identifying the critical aa residues of the known and unknown epitopes, and defining their roles in generating nAbs and non-nAbs will guide the rational design and construction of an efficacious EBV gp350-based vaccine that would focus B-cell responses to the protective epitopes.
In this study, we generated 23 hybridomas producing antibodies against gp350. To assess their clinical therapeutic/diagnostic potential and utility in informing future prophylactic vaccine design, we: (1) tested the ability of the new antibodies to detect gp350 protein, using enzyme-linked immunosorbent assay (ELISA), flow cytometry, and immunoblot; (2) sequenced the unique complementarity-determining regions (CDRs) of the heavy (VH) and light (VL) chains of all 23 hybridomas to identify novel mAbs; (3) measured the efficacy of each mAb to neutralize EBV infection of B cells in vitro; (4) used competitive cell binding assays to identify gp350 regions recognized by neutralizing and non-neutralizing mAbs and (5) used linear peptide binding assay to identify gp350 core linear aa residues recognized by neutralizing and non-neutralizing mAbs.
Results
Novel anti-gp350 mAbs target linear and conformational epitopes
Previous studies have generated and characterized several anti-gp350 mAbs, both neutralizing and non-neutralizing. Some of these have been effectively used to map the immunodominant or neutralizing epitopes present in the gp350 ectodomain, which has relevance for future strategies to design sterilizing prophylactic vaccines (Table 1). We generated a panel of new gp350-specific mAbs by immunizing BALB/c mice three times with purified UV-inactivated EBV. We further boosted the EBV-immunized mice three times with VLPs that incorporate the gp350 ectodomain (1–841) on the surface to improve antibody affinity maturation and avidity. We isolated splenocytes from the immunized mice and fused them with myeloma cells to generate hybridomas. We used indirect ELISA to screen supernatants from the hybridomas for specificity against purified gp350 ectodomain protein (aa 4–863) and identified 23 hybridomas producing gp350-specific antibodies (data not shown).
Table 1.
Summary of published gp350 epitope mapping using various methodologies
| Method | mAbs/protein/peptides | Number of epitopes identified | Reference |
|---|---|---|---|
| Competitive binding assay | mAbs | 7 epitopes – Sequence not defined | (Qualtiere et al., 1987a) |
| Binding studies: Determine the effects of anti-gp350 mAbs on gp350 binding to CR2 | mAbs | 2 possible regions identified by sequence alignment to C3d sequence: 1. aa 21–28 2. aa 372–378 |
(Nemerow et al., 1987a) |
| Peptide digest and immunoprecipitation | Truncated and mutant protein; mAbs (72A1 and BOS-1) | Narrowed down to the first 470 residues | (Tanner et al., 1988) |
| Binding studies | Peptide and protein | 2 sequences defined: 1. aa 21–28 2. N-terminus of gp350 |
(Nemerow et al., 1989) |
| Dot Blot immunoassay: Purified truncated protein incubated with mAbs | Protein – 8 clones overlapping N- and C-terminal portions of protein; mAbs from Qualtiere et al., 1987 | 3 sequences defined: 1. aa 310–325 2. aa 326–439 3. aa 733–841 |
(Zhang et al., 1991) |
| Peptide cell binding assay to 2 CR2-positive (Raji and Ramos) and 1 -negative (P3HR-1) cell lines | Synthesized peptides covering gp350 (907 aa) | 7 regions, 3 identified: 1. aa 142–161 2. aa 282–301 3. aa 822–841 |
(Urquiza et al., 2005) |
| Crystal structure and binding studies | Mutant proteins; mAbs 72A1 | 3 epitopes (based on 72A1 binding and gp350 4–443) 1. aa 16–29 2. aa 142–161 3. aa 282–301 |
(Szakonyi et al., 2006) |
| Structural docking studies and antigenicity mapping | gp350 and CR2 crystal structure alignment/docking | Single epitope (based on gp350 aa 1–470) 1. aa147–165 |
(Sitompul et al., 2012) |
| Structural alignment: computer modeling of gp350 and 72A1 and docking studies | Peptides (used in immunization); mAb (72A1) | 4 epitopes: identified 1. aa 14–20 2. aa 144–161 3. aa 194–211 4. aa 288–291 |
(Tanner et al., 2015) |
To further characterize the biochemical properties of the 23 antibodies generated, we purified the antibodies from the hybridoma supernatants using protein A spin columns, followed by SDS-PAGE to confirm the purity of all antibodies (Fig. 1A). When we reevaluated quantified amount of the purified antibodies (10 μg/ml) using indirect ELISA, all of the 23 antibodies and m72A1 (anti-gp350 positive control) had ELISA signals greater than two times those of anti-KSHV gH/gL mAb 54A1 (negative control), and were considered positive/specific to gp350 (Fig. 1B). Of the 23 gp350-positive hybridoma producing antibodies identified, HB4, HB5, HB7, HB13, and HB14 demonstrated binding strength equal to or greater than that of the positive control, m72A1. This difference in binding of the antibodies could be due to differential exposure of cognate epitopes on gp350 in the assay performed.
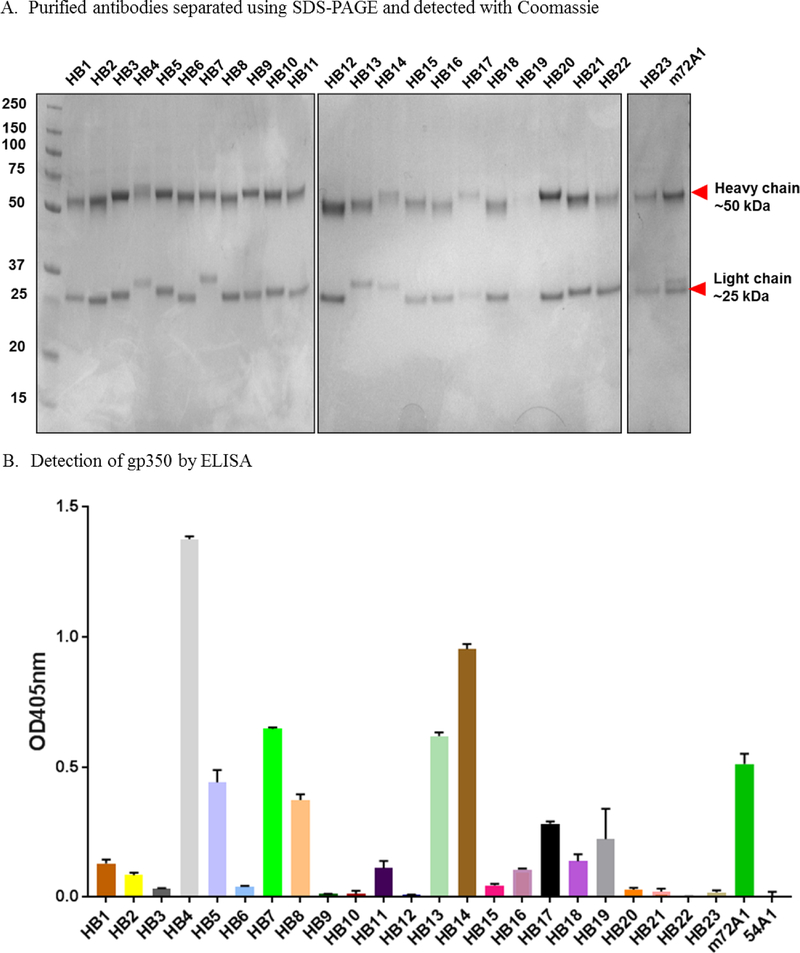
Figure 1. Specificity of anti-gp350 antibodies.
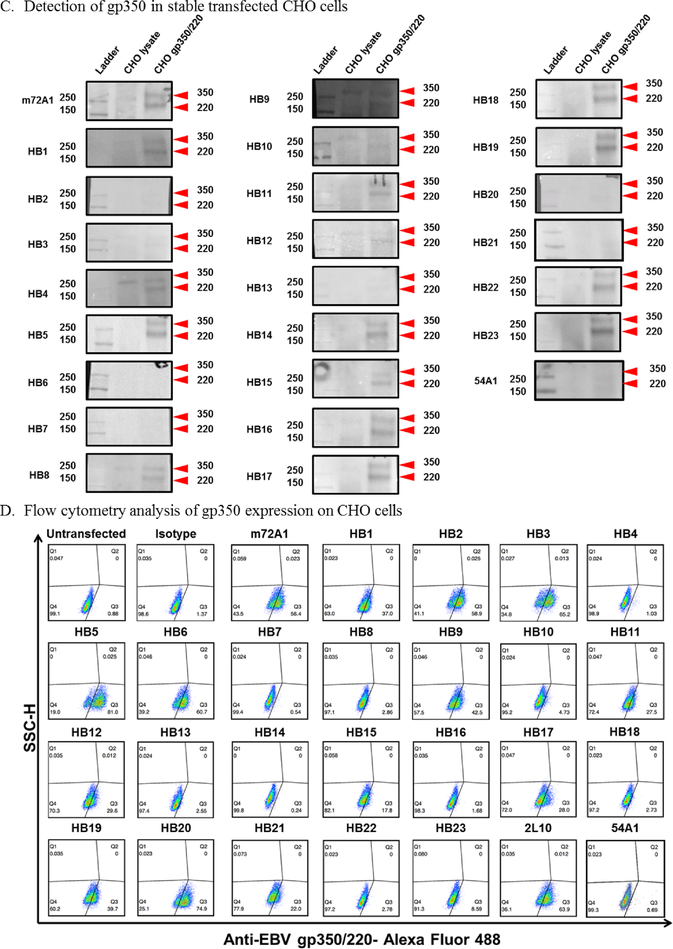
(A) SDS-PAGE analysis of anti-EBV gp350 antibodies purified from indicated hybridoma (HB) supernatants. (B) ELISA screening of HB supernatants for anti-gp350-specific antibodies. Soluble EBV gp350 protein was used as the target antigen at 0.5 μg/ml. m72A1 at 10 μg/ml and KSHV anti-gH/gl (54A1) were used as positive and negative (not shown) controls, respectively. Bound antibodies were detected using HRP-conjugated anti-mouse IgG (1:2,000). Twenty-three HB clones with ELISA signals two times greater than those of PBS control were considered to be positive/reactive to gp350. (C) Immunoblot analysis with gp350-transfected stable CHO lysate to determine specificity of anti-gp350-producing HB supernatants. (D) Flow cytometric analysis of surface expression of gp350 protein on gp350-expressing CHO cells. Cells were stained with indicated anti-gp350 mAbs (1:250), followed by secondary goat anti-mouse conjugated to AF488.
Determining the nature of the binding between an antibody and its target antigen is an important consideration for the performance and specificity of an antibody, as it can involve the recognition of linear or conformational epitopes (Sela et al., 1967). We evaluated the ability of the purified antibodies to bind linear epitopes by performing immunoblot analysis of denatured gp350 antigen expressed on CHO cells (Perez et al., 2017). We showed that 16 of the 23 antibodies detected both the 350 kDa and the 220 kDa splice variants (Fig. 1C). In contrast, HB2, HB3, HB6, HB7, HB13, HB20, and HB21, as well as the negative control 54A1, failed to recognize either of the denatured isoforms of gp350 (Fig. 1C). We further characterized the antibodies’ ability to bind conformational epitopes by using flow cytometric analysis of CHO cells stably expressing gp350 on the cell surface. HB1, HB2, HB3, HB5, HB6, HB9, HB11, HB12, HB15, HB17, HB19, HB20, HB21 and m72A1 readily recognized gp350 (Fig. 1D). The fact that HB2, HB3, HB6, HB20 and HB21 detected gp350 by flow cytometry, but not by immunoblot, suggests that these four antibodies only recognize conformational epitopes (i.e., native) on gp350, whereas HB5, HB9, HB11, HB15, HB17, and HB19 recognize epitopes present on both native and denatured linear protein (Fig. 1C–D). Importantly, the observation that all 23 anti-gp350 antibodies recognized the gp350 antigen either by indirect ELISA, immunoblot, or flow cytometry suggests that we successfully produced antibodies that are specific to EBV gp350 protein. In addition, we determined the isotypes of our newly generated antibodies to be IgG1 (n=14), IgG2a (n=5), IgG2b (n=1), a mixture of IgG1 and IgG2b (n=1), and a mixture of IgG1 and IgM (n=2) (Table 2).
Table 2.
Summarized biochemical and functional characterization of anti-gp350 antibodies
| Antibody | IgG sub-class | Light chain | ELISA binding to purified EBV gp350/220 | Flow cytometry (CHO Cells) | Western blot |
|---|---|---|---|---|---|
| HB1 | IgG1 | κ | + | + | + |
| HB2 | IgG2a | κ | + | + | − |
| HB3 | IgG2a | κ | + | + | − |
| HB4 | IgG1 | κ | + | − | + |
| HB5 | IgG2a | κ | + | + | + |
| HB6 | IgG1 | κ | + | + | − |
| HB7 | IgG1 | κ | + | − | − |
| HB8 | IgG1 | κ | + | − | + |
| HB9 | IgG2a | κ | + | + | + |
| HB10 | IgG1 | κ | + | − | + |
| HB11 | IgG1 | κ | + | + | + |
| HB12 | IgG1 | κ | + | + | + |
| HB13 | IgG1 | κ | + | − | − |
| HB14 | IgG1 | κ | + | − | + |
| HB15 | IgG1 | κ | + | + | + |
| HB16 | IgG1/ IgM | κ | + | − | + |
| HB17 | IgG2b | κ | + | + | + |
| HB19 | IgG1/ IgM | κ | + | + | + |
| HB20 | IgG2a | κ | + | + | − |
| HB21 | IgG1/IgG2b | κ | + | + | − |
| HB22 | IgG1 | κ | + | − | + |
| HB23 | IgG1 | κ | + | + | + |
| m72A1 | IgG1 | κ/ʎ | + | + | + |
Identification and characterization of 15 novel anti-350 monoclonal antibodies
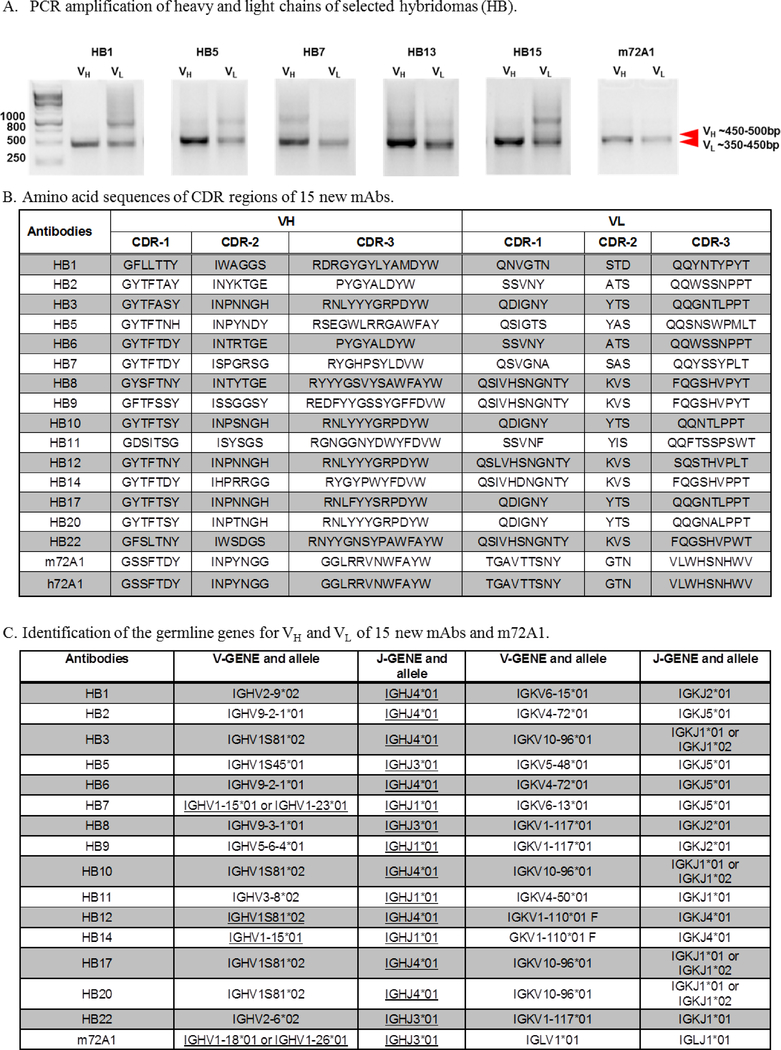
To determine whether the generated hybridomas were monoclonal or a mixture of antibodies, we sequenced the VH and VL variable region genes of the 23 new anti-gp350 antibodies, as well as m72A1 (positive control), using Illumina MiSeq. The sequence of the CDR of m72A1 antibody was recently determined and published (Herrman et al., 2015; Tanner et al., 2015). We used PCR to amplify the genes encoding the VH and VL chain regions in cDNAs generated from the 23 hybridoma cells, as well as from m72A1. The PCR products presented distinct bands for VH (~350–450 bp) and VL (~450–500 bp), Fig. 2A is a representation of identified VH and VL bands for only a few selected hybridomas.. We sequenced purified fragments, followed by in silico analysis, and identified CDRs for both VH and VL (Fig. 2B). As previously reported, we identified two unique IgG1 VH and two unique VL chains, one kappa and one lambda sequence of m72A1, using the light chain kappa degenerative primers and specific primers for the lambda light chain (Herrman et al., 2016). These sequences were >94% identical to the previously published sequences, suggesting that m72A1 exists as a mixture of antibodies, instead of the reported mAb (Tanner et al., 2015). Similar to m72A1, our HB4, HB13, HB15, and HB23 hybridomas each produced a mixture of two antibodies, with two unique sequences of the VH chain showing at >5% frequencies, suggesting that they are not mAbs (data not shown). We were unable to identify coding sequences for VL chains for HB7, HB9, and HB17, unless the frequencies were lowered to >1%; in this case, the identified coding VL chain sequences were identical.
Figure 2. Determination of novel anti-gp350 antibody sequences.
(A) Agarose gel analysis of PCR products of heavy chain of select novel anti-gp350 antibodies (HB1, HB4, HB7, HB13, and HB15) and m72A1 was used as a positive control. (B) Amino acid sequencing of the heavy (VH) and light (VL) chain variable region complementarity-determining regions (CDR) 1–3 of the new gp350 mAbs and mouse (m72A1) and humanized 72A1 (h72A1). (C) IMGT/V-QUEST analysis to determine the germline gene families for VH and VL of the 15 new mAbs and m72A1.
Our analysis and comparison of the VH and VL chain gene sequences of the 23 hybridomas compared to m72A1 showed unique sequences within the CDR 1–3 regions (Fig. 2B). Only HB8 and HB18 had identical VH and VL chain gene sequences, suggesting that the two are the same clone isolated separately; therefore, HB18 was excluded from subsequent experiments. One of the two paired sequences from the HB15 hybridoma mixture was confirmed to have identical VH and VL gene sequences to that of mAb HB10; however, based on our previous characterization, the presence of the additional paired sequence in the HB15 hybridoma was sufficient to confer subtle differences in biochemical interactions with gp350 between the HB10 and HB15 purified antibodies. In addition, we determined the germline genes for the VH and VL chains of the new 15 anti-gp350 mAbs and m72A1 (Fig. 2C). These results show that although only two mice were used in the generation of the antibodies, germline diversity was still present to some extent, and few mAbs shared the same germline gene rearrangement and evolution. Thus, sequence analysis demonstrated that we generated 15 unique anti-gp350 mAbs, with distinct sequence identities from the commercially available m72A1. The sequence of the widely used non-neutralizing antibody 2L10 (originally from G. Pearson’s laboratory) is not currently available and we were not able to access the hybridoma; thus, 2L10 was not used in our sequence comparative studies.
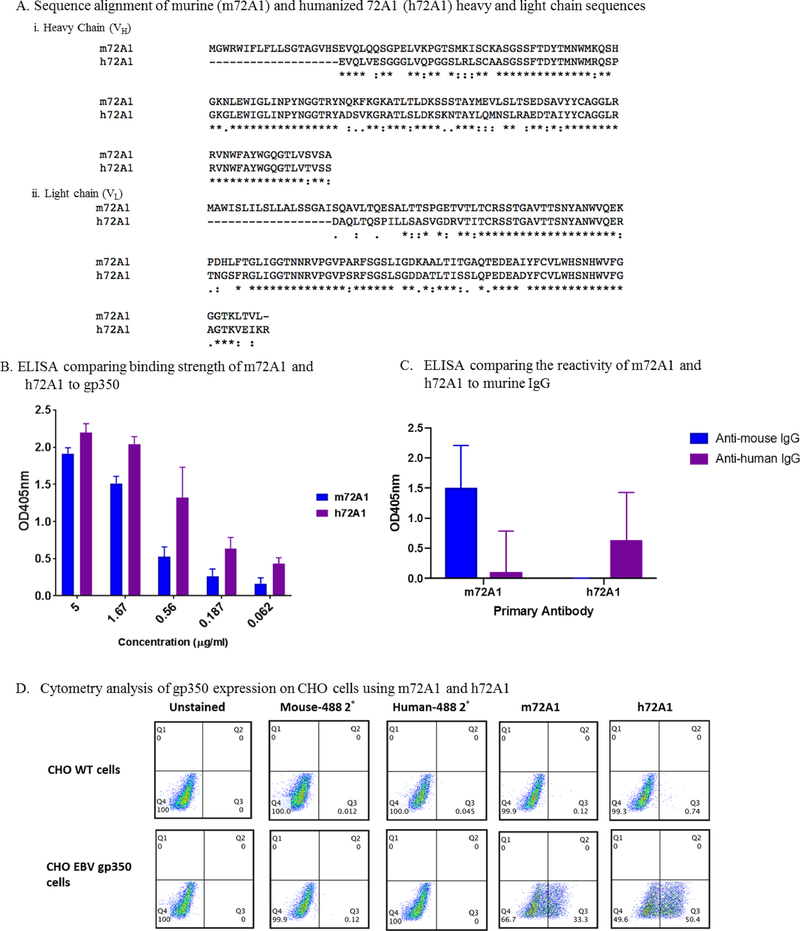
Humanization of m72A1 abrogated murine immunogenicity
The m72A1 VH and VL chain sequences identified in this study were identical to the ones published by Herrman et al., and we used them to generate a humanized 72A1 (h72A1) as a strategy to reduce and/or eliminate human anti-murine antibody (HAMA) (Fig. 3A). Our h72A1 bound gp350 with similar strength to m72A1 in ELISA (Fig. 3B). We further determined the levels of anti-mouse and anti-human activity retained in our h72A1 nAb using ELISA. As shown in Fig. 3C, mouse 72A1 mAb reacted strongly to goat-anti-mouse IgG as compared to goat anti-human IgG (9-fold, p< 0.0001) and 28-fold above the background (1x PBS) (p< 0.0001). In contrast, h72A1 mAb did not react at all to goat anti-mouse, but specifically reacted strongly to goat anti-human IgG (2100-fold, p <0.0001) over the background. To determine whether h72A1 still recognized gp350 in its native conformation, we performed flow cytometric analysis. We showed that h72A1 recognized native epitopes of gp350 expressed on CHO cells surface, comparable to m72A1 (Fig. 3D). These results indicate that humanization of m72A1 did not affect its ability to recognize native gp350, but it abrogated anti-mouse reactivity and increased anti-human reactivity.
Figure 3. Comparison of murine 72A1 (m72A1) and humanized 72A1 (h72A1).
(A) Sequence comparison of murine (m72A1) and humanized (h72A1) 72A1. ClustalW alignment of heavy chain (i) and light chain (ii) variable region amino acid sequences. Regions of identical sequence are represented by *. Regions of similarity are represented by:. (B) ELISA comparison screening of m72A1 and h72A1 for anti-gp350-specificity. Soluble EBV gp350 protein was used as the target antigen at 0.5 μg/ml. m72A1 and h72A1 were serially diluted (5–0.062 μg/ml) and 1x phosphate buffered saline (PBS) was used as a negative control (data not shown). Bound h72A1 and m72A1 antibodies were detected using HRP-conjugated anti-mouse IgG and anti-human IgG (1:2,000) as relevant. (C) ELISA determining the reactivity of humanized 72A1 to murine IgG. Soluble EBV gp350 protein was used as the target antigen at 0.5 μg/ml. Plates were incubated with 10 μg/ml of m72A1 and h72A1, followed by three washes. Bound antibodies were detected using HRP-conjugated anti-mouse IgG or anti-human IgG (1:2,000). (D) Flow cytometric analysis of m72A1 and h72A1 gp350 specificity. CHO wild-type cells and gp350-expressing CHO cells were stained with m72A1 and h72A1, followed by secondary goat anti-mouse or anti-human conjugated to AF488. Unstained cells and cells stained with secondary goat anti-mouse or anti-human conjugated to AF488 alone were used as negative controls. 2° represents secondary antibody.
Novel anti-gp350 HB5 neutralizes EBV infection to B cells at comparable levels to that of m72A1 and h72A1 antibodies
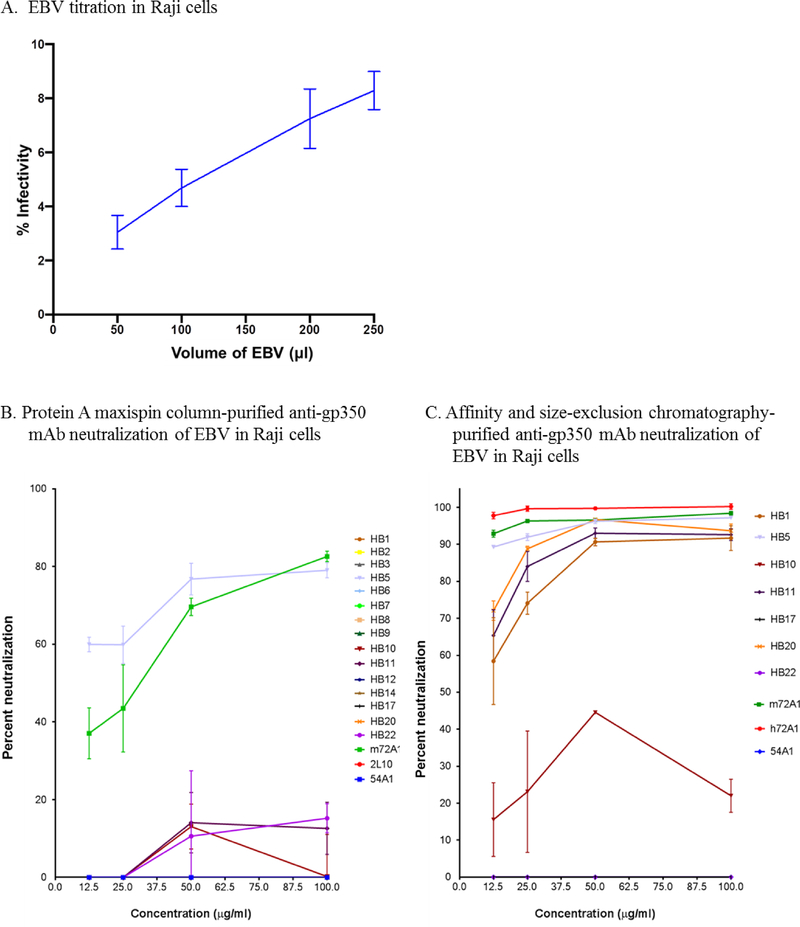
Currently, m72A1 is the only commercially available anti-gp350 nAb (Hoffman et al., 1980). However, this antibody was recently reported to be a mixture of a gp350-specific and MOP-specific antibodies (Herrman et al., 2016). We evaluated the ability of our 15 newly identified mAbs to neutralize purified eGFP-tagged-EBV infection of a B-cell line (Raji) in vitro compared to that of m72A1 (mixture) and our newly cloned and biochemically characterized h72A1, following standardized procedures (Ogembo et al., 2015; Sashihara et al., 2009). We determined the percentage of eGFP+ cells (percent infection) using flow cytometry, as described (Ogembo et al., 2015). Anti-gp350 non-neutralizing mAb 2L10 and anti-KSHV gH/gL mAb 54A1 antibodies were used as negative controls. Because HB4, HB13, HB15, HB16, HB19, HB21, and HB23 were confirmed to be polyclonal based on isotyping and/or sequence data, we eliminated them from further consideration in the neutralization assay. HB18 was not used in neutralization experiments because it was identical to HB8. First, we titrated purified eGFP-tagged-EBV in Raji cells to determine percent EBV infection using a range of volumes (50–250 μl) (Fig. 4A). Then we conducted initial neutralization of EBV in Raji cells using purified mAbs at various concentrations (12.5, 25, 50, and 100 μg/ml). Only HB1, HB5, HB11, HB22 and m72A1 showed a dose-dependent neutralization of EBV in Raji cells; the neutralization capability of HB5 (60–80%) was comparable to that of m72A1 (35–80%) (Fig. 4B). In contrast, HB2, HB3, HB6, HB7, HB8, HB9, HB10, HB12, HB14, HB17, and HB20 mAbs failed to neutralize EBV infection, even at the highest concentration. As expected, neither 2L10 nor 54A1 neutralized EBV infection, even at the highest mAb concentration of 100 μg/ml (Fig. 4B).
Figure 4: Neutralization activity of novel anti-gp350 mAbs against EBV-eGFP in Raji cells.
(A) EBV-eGFP titration in Raji cells to determine optimal dose of infection. (B) EBV-eGFP was pre-incubated with 15 indicated serial diluted (12.5–100 μg/ml), maxispin column-purified anti-gp350 mAbs, followed by incubation with 105 Raji cells for 48 h. EBV-eGFP+ cells were enumerated using flow cytometry. Anti-gp350 (m72A1) nAb served as positive control and non-neutralizing anti-gp350 (2L10) mAb and anti-KSHV gH/gL mAb (54A1) served as negative controls. (C) EBV-eGFP was pre-incubated with 7 indicated serially diluted (12.5–100 μg/ml) protein G affinity chromatography- and size-exclusion chromatography-purified anti-gp350 mAbs, followed by incubation with 105 Raji cells for 48 h. EBV-eGFP+ cells were enumerated using flow cytometry. Anti-gp350 (m72A1 and h72A1) nAbs served as positive controls and anti-KSHV gH/gL mAb (54A1) served as negative control.
Subsequently, we further purified seven representative novel nAb and non-nAb anti-gp350 mAbs (HB1, HB5, HB10, HB11, HB17, HB20 and HB22) as well as controls (m72A1, h72A1 and 54A1) using protein G affinity chromatography and size-exclusion chromatography in order to eliminate any potential impurities, then reevaluated their potency in blocking EBV infection of Raji cells. Chromatography-purified HB1, HB5, HB11, and HB20, blocked EBV infection in a dose-dependent manner (Fig. 4C). HB5 was the most effective nAb among newly generated mAbs in our laboratory, efficiently blocking EBV infection (90%) at percentages comparable to both m72A1 (93%) and h72A1 (98%), even at the lowest concentration of 12.5 μg/ml, with 97% nAb activity at 100 μg/ml (Fig. 4C). HB1, HB11, and HB20 neutralized EBV infection between 57–73% at the lowest concentration (12.5 μg/ml) and 90% at 100 μg/ml. Neither HB17, HB22, nor 54A1 blocked EBV infection; although HB10 blocked some EBV infection, nAb activity did not reach 50% even at the highest concentration of antibody used, thus we also classified it as a non-nAb.
Four novel gp350 nAbs bind antigenic epitopes that overlap with those of 72A1
At least seven unique CD21 binding epitopes on EBV gp350 have been predicted (Table 1). One of these epitopes (aa 142–161) has been identified as the primary epitope recognized by m72A1 (Szakonyi et al., 2006) and mice immunized with the 142–161 peptide elicit nAbs against EBV infection (Tanner et al., 2015). To evaluate whether the selected novel nAbs (HB1, HB5, HB11, and HB20) and non-nAbs (HB10, HB17, and HB22) bind overlapping or non-overlapping target epitopes to those of 72A1, we determined their ability to compete for binding to gp350 expressed stably on transfected CHO cells. We observed antigen binding competition between biotinylated m72A1 (1 μg/ml) and serially diluted (500, 250, 125, and 67.5 μg/ml) unlabeled gp350 nAbs (HB1, HB5, HB11, HB20, and h72A1), but not the gp350 non-nAbs (HB10, HB17, or HB22) or anti-KSHV gH/gL antibody 54A1 (negative control) (Table 3). However, previously non-nAbs HB10 and HB22, were shown not to bind native gp350 expressed on CHO cells using FACS (Fig 1D), suggesting that the observed lack of competitive binding could be attributed to these two Abs not binding the native gp350 expressed on the CHO cells. These results indicate that nAbs HB1, HB5, HB11, and HB20, as well as h72A1, bind overlapping target epitopes with that of m72A1, while non-nAbs HB17, the only non-nAbs able to recognize gp350 in conformational form, binds different target epitopes. We obtained similar results when we performed cross-competition binding assays between 1 μg/ml biotinylated and 500 μg/ml unlabeled gp350 nAbs HB1, HB5, HB11, HB20, m72A1, and h72A1 and non-nAb HB17 (Table 4), confirming that the newly developed gp350 nAbs bind overlapping epitopes to 72A1. Because of inability of non-nAbs, HB10 and HB22 to recognize conformational gp350, they were excluded from the cross competitive cell binding assays. Importantly, even though HB1, HB5, HB11, and HB20 competed with 72A1 for the same antigenic epitope, each of these nAbs had unique VH and VL sequences from 72A1 (Fig. 2B).
Table 3.
Cell binding mAbs competition assay with EBV gp350 using biotinylated m72A1
| Unlabeled mAbs | % inhibition of biotinylated nAbs | |||
|---|---|---|---|---|
| 500 μg/ml | 250 μg/ml | 125 μg/ml | 67.5 μg/ml | |
| HB1 | 91 | 89 | 81 | 81 |
| HB5 | 95 | 93 | 92 | 94 |
| HB10 | 12 | 14 | 16 | 13 |
| HB11 | 95 | 95 | 89 | 87 |
| HB17 | 32 | 32 | 6 | 0 |
| HB20 | 96 | 91 | 87 | 89 |
| HB22 | 10 | 3 | 16 | 9 |
| m72A1 | 91 | 93 | 94 | 97 |
| h72A1 | 98 | 95 | 93 | 89 |
| 54A1 | 18 | 7 | 10 | 4 |
Table 4.
Cross-competitive binding of EBV gp350
| % inhibition of biotinylated nAbs | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Unlabeled mAbs | HB1 | HB5 | HB10 | HB11 | HB17 | HB20 | HB22 | m72A1 | h72A1 | 54A1 |
| HB1 | 92 | 97 | ND | 96 | 16 | 61 | ND | 98 | 95 | 4 |
| HB5 | 92 | 98 | ND | 93 | 9 | 73 | ND | 97 | 97 | 13 |
| HB10 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| HB11 | 93 | 96 | ND | 96 | 17 | 3 | ND | 97 | 97 | 9 |
| HB17 | 23 | 32 | ND | 11 | 86 | 17 | ND | 24 | 16 | 0 |
| HB20 | 88 | 97 | ND | 89 | 13 | 59 | ND | 98 | 96 | 0 |
| HB22 | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| m72A1 | 82 | 97 | ND | 90 | 37 | 31 | ND | 98 | 98 | 1 |
| h72A1 | 87 | 97 | ND | 79 | 31 | 26 | ND | 98 | 98 | 0 |
| 54A1 | 0 | 0 | ND | 0 | 0 | 0 | ND | 0 | 3 | 0 |
ND- Not determined
Novel anti-gp350 nAbs and non-nAbs bind three major immunodominant regions on gp350
The gp350 ectodomain is heavily glycosylated, with both N- and O-linked sugars, which accounts for more than half of the 350-kDa molecular mass of the protein. Currently, there is only one crystal structure available for gp350, comprising a truncated structure between 4–443 aa, with at least 14 glycosylated asparagine residues coating the protein with sugars, with the exception of a single glycan-free patch (Szakonyi et al., 2006). Mutation of several residues in the glycan-free patch resulted in the loss of CD21 binding (Szakonyi et al., 2006), suggesting that binding of CD21 by gp350 is mediated within this region. To identify linear epitopes on gp350, we scanned anti-gp350 nAbs (HB1, HB5, HB11 and HB20) and non-nAbs (HB10, HB17, and HB22) in an ELISA-based assay using a peptide library consisting of sequential peptides (Supplementary Fig. 1). The peptide library, consisting of 20-mer peptides, covered the entire gp350 protein B95–8 sequence (Baer et al., 1984), with the exception of aa 862–881 (Table 5). This peptide could not be synthesized due to high hydrophobicity of aa residues in the sequence. We used purified m72A1 and h72A1 nAbs as positive controls and anti-KSHV gH/gL 54A1 antibody as a negative control. We used purified recombinant gp350 ectodomain as a control to validate the binding activity for all of the antibodies used. As shown in Supplementary Fig. 1, the binding strength and binding pattern of the mAbs to the linear peptides varied, although similarities could be observed between non-nAbs, as well as nAbs. All newly generated nAbs as well as h72A1 and m72A1 bound to peptide 8, which was composed of the sequence 142-HHAEMQNPVYLIPETVPYIK-161, with the exception of HB1. This aa sequence has previously been identified to be part of the neutralizing epitope on gp350, as well as a binding site for nAb, 72A1. As expected, strong binding was observed for mAbs to purified recombinant gp350 ectodomain, with the exception of HB22.
Table 5.
Sequence, length and position of EBV gp350 peptides
| Peptide name | Peptide | gp350 region | Peptide sequence | Peptide length |
|---|---|---|---|---|
| Pep-1 | 1–20 | 1 | MEAALLVCQYTIQSLIHLTG | 20 |
| Pep-2 | 21–40 | 1 | EDPGFFNVEIPEFPFYPTCN | 20 |
| Pep-3 | 41–61 | 1 | VCTADVNVTINFDVGGKKHQL | 21 |
| Pep-4 | 62–81 | 1 | DLDFGQLTPHTKAVYQPRGA | 20 |
| Pep-5 | 82–101 | 1 | FGGSENATNLFLLELLGAGE | 20 |
| Pep-6 | 102–121 | 2 | LALTMRSKKLPINVTTGEEQ | 20 |
| Pep-7 | 122–141 | 2 | QVSLESVDVYFQDVFGTMWC | 20 |
| Pep-8 | 142–161 | 2 | HHAEMQNPVYLIPETVPYIK | 20 |
| Pep-9 | 162–181 | 2 | WDNCNSTNITAVVRAQGLDV | 20 |
| Pep-10 | 182–201 | 2 | TLPLSLPTSAQDSNFSVKTE | 20 |
| Pep-11 | 202–221 | 3 | MLGNEIDIECIMEDGEISQV | 20 |
| Pep-12 | 222–241 | 3 | LPGDNKFNITCSGYESHVPS | 20 |
| Pep-13 | 242–261 | 3 | GGILTSTSPVATPIPGTGYA | 20 |
| Pep-14 | 262–281 | 3 | YSLRLTPRPVSRFLGNNSIL | 20 |
| Pep-15 | 282–301 | 3 | YVFYSGNGPKASGGDYCIQS | 20 |
| Pep-16 | 302–321 | 4 | NIVFSDEIPASQDMPTNTTD | 20 |
| Pep-17 | 322–341 | 4 | ITYVGDNATYSVPMVTSEDA | 20 |
| Pep-18 | 342–361 | 4 | NSPNVTVTAFWAWPNNTETD | 20 |
| Pep-19 | 362–381 | 4 | FKCKWTLTSGTPSGCENISG | 20 |
| Pep-20 | 382–401 | 4 | AFASNRTFDITVSGLGTAPK | 20 |
| Pep-21 | 402–421 | 5 | TLIITRTATNATTTTHKVIF | 20 |
| Pep-22 | 422–441 | 5 | SKAPESTTTSPTLNTTGFAD | 20 |
| Pep-23 | 442–461 | 5 | PNTTTGLPSSTHVPTNLTAP | 20 |
| Pep-24 | 462–481 | 5 | ASTGPTVSTADVTSPTPAGT | 20 |
| Pep-25 | 482–501 | 5 | TSGASPVTPSPSPWDNGTES | 20 |
| Pep-26 | 502–521 | 6 | KAPDMTSSTSPVTTPTPNAT | 20 |
| Pep-27 | 522–541 | 6 | SPTPAVTTPTP NATSPTPAV | 20 |
| Pep-28 | 542–561 | 6 | TTPTPNATSPTLGKTSPTSA | 20 |
| Pep-29 | 562–581 | 6 | VTTPTPNATSPTLGKTSPTS | 20 |
| Pep-30 | 582–601 | 6 | AVTTPTPNATSPTLGKTSPT | 20 |
| Pep-31 | 602–621 | 7 | SAVTTPTPNATGPTVGETSP | 20 |
| Pep-32 | 622–641 | 7 | QANATNHTLGGTSPTPVVTS | 20 |
| Pep-33 | 642–661 | 7 | QPKNATSAVTTGQHNITSSS | 20 |
| Pep-34 | 662–681 | 7 | TSSMSLRPSSNPETLSPSTS | 20 |
| Pep-35 | 682–701 | 7 | DNSTSHMPLLTSAHPTGGEN | 20 |
| Pep-36 | 702–721 | 8 | ITQVTPASISTHHVSTSSPA | 20 |
| Pep-37 | 722–741 | 8 | PRPGTTSQASGPGNSSTSTK | 20 |
| Pep-38 | 742–761 | 8 | PGEVNVTKGTPPQNATSPQA | 20 |
| Pep-39 | 762–781 | 8 | PSGQKTAVPTVTSTGGKANS | 20 |
| Pep-40 | 782–801 | 8 | TTGGKHTTGHGARTSTEPTT | 20 |
| Pep-41 | 802–821 | 9 | DYGGDSTTPRPRYNATTYLP | 20 |
| Pep-42 | 822–841 | 9 | PSTSSKLRPRWTFTSPPVTT | 20 |
| Pep-43 | 842–861 | 9 | AQATVPVPPTSQPRFSNLSM | 20 |
| Pep-44 | 862–881 | 9 | LVLQWASLAVLTLLLLLVMA | 20 |
| Pep-45 | 882–901 | 9 | DCAFRRNLSTSHTYTTPPYD | 20 |
| Pep-46 | 888–907 | 9 | NLSTSHTYTTPPYDDAETYV | 20 |
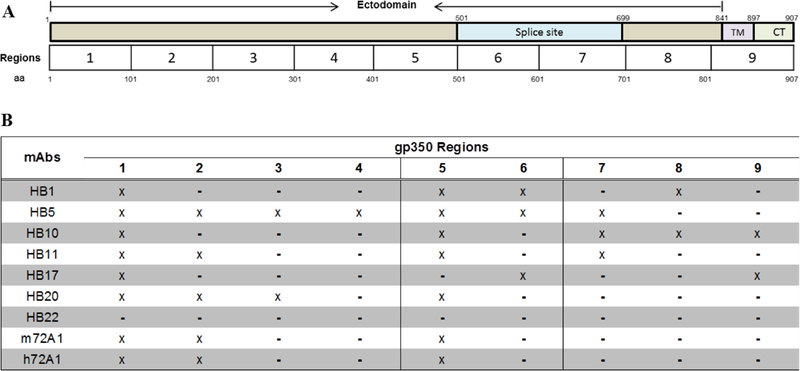
We divided the overall gp350 sequence into nine different regions consisting of ~100 aa (Fig. 5A). The three major regions that exhibited the greatest affinity to anti-gp350 mAbs were: 1–101, 102–201, and 402–501 (Fig. 5B). The aa 102–201 region was bound by only nAbs (HB5, HB11, HB20, m72A1 and h72A1), with the exception of HB1. Notably, this region (102–201) contains the epitope (aa 142–161) previously identified as a binding epitope for 72A1 and as a binding receptor for CR2 (Table 1), confirming that this is the main region that interacts with most gp350 nAbs. Because both nAbs and non-nAbs bound to aa 1–101 and 402–501, we considered these two regions to be immunodominant.
Figure 5. Analysis of anti-gp350 antibodies linear epitope binding.
(A) Schematic diagram of EBV gp350 protein, illustrating the ectodomain and the splice site (aa 501–699) for making gp220, the transmembrane domain, TM (aa 841–897) and the cytoplasmic domain, CT (aa 898–907). To analyze and classify binding of anti-gp350 mAbs to linear epitopes on the protein, EBV gp350 was separated into 9 regions of ~100 aa. (B) Summarized analysis of anti-gp350 mAb linear epitope binding to various regions of gp350. Three major immunodominant regions were identified, region 1 (aa 1–100), 2 (aa 101–202) and 5 (aa 401–502).
Discussion
Antibodies, whether elicited in the host naturally or via passive immunization, provide an effective first-line of defense and correlate with protection against viral infection in humans (Iwasaki, 2016). In the past five decades, several immunization studies have indicated that the EBV major immunodominant gp350 is an ideal target for eliciting nAbs in immunized animals and humans (Cohen, 2015, 2018; Cohen et al., 2011; Cohen et al., 2013). Although the ectodomain of gp350 (aa 1–841) contains at least seven unique CD21 binding epitopes (Table 1), only one of these epitopes (aa 142–161) has been shown to be capable of eliciting nAbs (Hoffman et al., 1980; Szakonyi et al., 2006; Tanner et al., 1988; Tanner et al., 2015; Urquiza et al., 2005). The role of the remaining six predicted epitopes in generating nAbs has not been elucidated; however, this information is needed to justify the recent use of only a small fragment of gp350 in the design of EBV prophylactic vaccines (Cui et al., 2013; Kanekiyo et al., 2015).
In this study, we generated 23 hybridomas producing gp350-specific antibodies and biochemically characterized their ability to bind gp350. Out of the 23 hybridomas, we determined that 15 were monoclonal and novel, based on their VH and VL CDR sequences, compared to the reported sequence of m72A1 (Herrman et al., 2016). Following confirmation that the new 15 mAbs recognized gp350 antigen and contained unique VH–VL sequences, further characterization revealed that mAbs HB1, HB5, HB11, and HB20 inhibited EBV infection of a human B-cell line in a dose-dependent manner, with HB5 being the best neutralizer, comparable to m72A1 and h72A1. Thus, our study provides four new nAbs against EBV infection of B cells with potential clinical utility in blocking viral infection in immunosuppression settings.
For patients with end-stage organ failure or hematologic malignancies, transplantation is the treatment of choice (Rickinson and Kieff, 2007a). However, transplantation success depends entirely on use of potent immunosuppressive drugs to prevent stem cell/organ rejection, which in turn can impose serious side effects, including increased risk of infection with or reactivation of EBV, and resultant development of EBV+ post-transplant lymphoproliferative diseases (EBV+ PTLDs) and other types of lymphomas. EBV+ PTLDs are aggressive, life-threatening complications, especially for pediatric transplant recipients who are EBV-naïve pre-transplantation (Coté et al., 1998; Goedert et al., 1998). A variety of non-standardized, non-specific treatments are currently used to treat EBV+ PTLD cases with variable success (Benkerrou et al., 1998; Cruz et al., 2012; Faro et al., 1996; Milpied et al., 2000; Papadopoulos et al., 1994; Starzl and Holmes, 1964). Currently, non-Hodgkin lymphoma and other EBV-associated B-cell lymphomas are the most common PTLDs seen in the pediatric transplant population (Engels et al., 2011; Yanik et al., 2017). Thus, the need for novel EBV-specific immunotherapies that could potentially block infection and/or target EBV+ cells to prevent PTLDS and/or other lymphomas are needed.
The recent identification and isolation of nAbs and their use in the clinic to prevent infection against highly variable viruses (e.g., HIV-1 (Stamatatos et al., 2009; Walker et al., 2009), influenza (Han and Marasco, 2011; Sui et al., 2009; Wrammert et al., 2008), respiratory syncytial virus (Piedimonte et al., 2000), and cytomegalovirus (Alexander et al., 2010)), suggests that a similar strategy could be successful in prevention of EBV infection. In 2012, an international, multidisciplinary expert panel (“The Seville expert workshop for progress in PTLDs”) recommended use of intravenous anti-viral nAbs for preventing or treating EBV+ PTLDs (Glotz et al., 2012). Thus, nAbs offer a promising approach for EBV therapeutic vaccines, once they are deemed safe and potent enough. In a small phase I clinical trial, m72A1 conferred short-term protection against acquiring EBV after transplantation in 3 of 4 pediatric patients (Haque et al., 2006). However, there was a major drawback: all 4 patients who received m72A1 developed human anti-mouse antibodies (HAMA), which caused side effects and limited treatment efficacy, and one patient developed a hypersensitivity reaction. This suggests that m72A1 is not safe for humans; generation of chimeric (human/mouse) or humanized 72A1 nAbs is expected to overcome safety complications associated with HAMA. In addition, EBV uses multiple envelope glycoproteins, including the immunodominant gp350 and the gH/gL complex, to infect host cells (Connolly et al., 2011; Eisenberg et al., 2012). These glycoproteins are expressed on both EBV virions and lytically induced EBV+ cells (Cohen et al., 2011; Henle et al., 1968), and stimulate immune responses in humans and in animal models (Khanna et al., 1999; Perez et al., 2016; Thorley-Lawson and Geilinger, 1980), making them attractive combination targets for a potentiated EBV nAb therapeutic vaccine (Cohen, 2015). Indeed, our recent pre-clinical studies showed that sera from mice immunized with both gp350 and gH/gL vaccines prevented EBV infection better than individual immunogens (Perez et al., 2016). Other groups showed that mAbs against gp350 (m72A1) (Ogembo et al., 2013; Ogembo et al., 2015; Thorley-Lawson and Geilinger, 1980) and anti-gH/gL human mAbs (AMMO1) (Snijder et al., 2018) or (769B10) (Bu et al., 2019) blocked in vitro EBV infection of B cells and epithelial cells, respectively, the two primary cell types targeted by the virus in vivo. Targeting all glycoproteins important for infection of various cell types will improve the potency of the nAbs as a strategy of preventing EBV infection.
Our identification of four new potent nAbs (HB1, HB5, HB11 and HB20) and sequencing of their CDR regions, as well as our humanization of m72A1, opens the possibility of using these nAbs for clinical applications, such as reducing or preventing EBV infection in transplant settings, with the consequent potential to reduce the incidence of EBV+ PTLDs. Our h72A1 IgG1 antibody recognized both native gp350 as well as gp350 peptides that constitute the principal gp350 neutralizing epitope (142–161) and completely eliminated anti-murine IgG immunoreactivity. Importantly, h72A1and our four newly generated nAbs (HB1, HB5, HB11, and HB20)—significantly blocked in vitro EBV infection of B cells to a degree comparable to or better than m72A1. These results are consistent with recent data from chimeric and/or humanized 72A1 neutralization assay in Raji cells (Tanner et al., 2018). Ongoing experiments in our laboratory are focused on humanizing the four potent nAbs, followed by comparative studies with h72A1 and combination use with anti-gH/gL humanized and/or human antibodies, in which we will assess their ability to block infection of both epithelial and B cells in vitro and in a humanized mouse model. We expect that combining nAbs that bind to different peptides on gp350 and gH/gL will significantly reduce infection in both B cells and epithelial cells.
In addition, both nAbs and non-nAbs can be used as research tools to provide insight into epitope targets important for vaccine development. In the past, various methods, including lectin/ricin immune-affinity assay (Thorley-Lawson and Geilinger, 1980), purified mAbs (Alfarano et al., 2005; Qualtiere et al., 1987b), purified soluble gp350 mutants, synthetic peptides (Nemerow et al., 1989; Nemerow et al., 1987b; Tanner et al., 2015), cell binding assays (Urquiza et al., 2005), and X-ray crystallography of partial gp350 protein (aa 4–443) (Szakonyi et al., 2006), have been used to identify the critical gp350 epitopes responsible for its interaction with the CD21 and CD35 cellular receptors (summarized in Table 1). Despite several attempts to identify gp350 epitopes important for eliciting nAbs, to date only a single epitope, aa 142–161, has been identified, which is also the binding epitope for nAb 72A1. Currently, the lack of a crystal structure of full-length gp350 protein and the unavailability of multiple nAbs hinder the opportunity to identify other gp350 epitopes that might elicit nAbs and inform design of effective vaccine strategies. To identify gp350 epitopes responsible for eliciting nAbs, we took advantage of our newly generated four nAbs (HB1, HB5, HB11, and HB20) and three non-nAbs (HB10, HB17, and HB22) to perform competitive cell binding and ELISA-based linear peptide binding assays. Although both approaches have various limitations, they offer useful information that when combined might inform and/or advance vaccine development efforts. Competitive cell binding assays can provide information on whether two antibodies bind overlapping or non-overlapping epitopes, although they are unable to indicate whether the competing antibodies bind the same or nearby epitopes, nor identify actual aa residues involved in the binding (Ladner, 2007). On the other hand, the ELISA peptide binding assay is only reactive to linear epitopes and may or may not take into consideration post-translational protein modifications, depending on whether a full protein or peptides are used as the target antigen(s).
Using biotinylated antibodies, we showed that the newly generated gp350 nAbs (HB1, HB5, HB11, and HB20) bound targets that overlapped with those of both m72A1 and h72A1, although HB20 showed only partial binding to the overlapping targets. The non-Ab, HB17 showed little to no competitive binding when compared to nAbs, suggesting that they bound different gp350 epitopes. These results strongly suggest that we have identified two distinct binding regions, one bound predominantly by nAbs and the other by non-nAbs, and that nAbs potentially bind targets within the same proximity, if not the same aa sequences. Thus, the current antibodies provide the first step toward generating reagents required for mapping neutralizing versus non-neutralizing epitopes on gp350, should the full-length crystal structure of the protein remain unavailable. Using linear peptide epitope mapping, we identified three major mAb-binding regions, 1–101, 102–201, and 402–501; all three regions incorporable previously identified epitopes (Tanner et al., 2015; Urquiza et al., 2005; Zhang et al., 1991). Regions 1–101 and 402–501 were bound by both nAbs and non-nAbs, suggesting that these regions are immunodominant. However, the 102–201 region containing the nAb epitope 142–161 was only bound by nAbs (HB5, HB11, HB20, and both m72A1 and h72A1), with the exception of the nAb HB1. We expect further studies involving alanine scanning of the positive peptides to elucidate the exact aa residues of the linear epitopes bound by nAbs. These results suggest that epitopes/regions capable of eliciting nAbs are located within the N-terminus of gp350.
In conclusion, we generated 15 novel anti-gp350-specific mAbs, characterized their binding to gp350, determined their neutralization activity against EBV infection in vitro, mapped their cognate epitopes, and defined the linear epitopes they recognize on gp350 aa residues. We identified 3 major regions responsible for generating nAbs and non-nAbs and narrowed down their binding epitopes to a region of ~100 aa residues. This study also provides additional evidence to support the current usage of the N-terminus of gp350 aa 4–443 (Szakonyi et al., 2006) as a vaccine candidate, due to the presence of the antigenic epitope(s) capable of eliciting nAbs.
Materials and Methods
Cells and viruses
AGS-EBV-eGFP, a human gastric carcinoma cell line infected with a recombinant Akata virus expressing enhanced green fluorescent protein (eGFP) was a kind gift of Dr. Liisa Selin (University of Massachusetts Medical School). Chinese hamster ovary cells (CHO); human embryonic kidney cells expressing SV-40 T antigen (HEK-293T); HEK-293 6E suspension cells; EBV-positive Burkitt lymphoma cells (Raji); myeloma cells (P3X63Ag8.653); and anti-EBV gp350 m72A1 hybridoma cells (HB168) were purchased from American Type Culture Collection (ATCC). ExpiCHO cells were purchased from ThermoFisher Scientific.
AGS-EBV-eGFP cells were maintained in Ham’s F-12 media supplemented with 500 μg/ml neomycin (G418, Gibco). Raji, P3X63Ag8.653, and HB168 hybridoma cells were maintained in RPMI 1640. CHO and HEK-293T cells were maintained in DMEM. HEK-293 6E and ExpiCHO cells were maintained in FreeStyle F17 Expression Medium supplemented with 0.1% Pluronic F-68 and Gibco ExpiCHO Expression Media, respectively. All culture media were supplemented with 10% fetal bovine serum (FBS) from Millipore Sigma, 2% penicillin-streptomycin, and 1% l-glutamine, with the exception of Freestyle F17 expression and Gibco ExpiCHO Expression Media. All media were purchased from ThermoFisher Scientific unless otherwise specified.
Antibodies and plasmids
Primary antibodies: EBV gp350 nAb (m72A1) was purified from the supernatant of the HB168 hybridoma cell line using Capturem™ Protein A Maxiprep spin columns (Takara) or protein G affinity and size-exclusion chromatography. The non-nAb gp350/220 mAb (2L10) was purchased from Millipore Sigma. Anti-KSHV gH/gL 54A1 mAb was generated and characterized in our laboratory as outlined (Mulama et al., 2019).
Secondary antibodies: Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG for immunoblot or ELISA was purchased from Bio-Rad. HRP-conjugated goat anti-human IgG for ELISA was purchased from ThermoFisher Scientific. Alexa Fluor® (AF) 488-conjugated goat anti-mouse IgG (H+L) and AF488-conjugated goat anti-human IgG (H+L) for flow cytometry was purchased from Life Sciences Tech. Anti-biotin monoclonal antibody conjugated to AF488 for competitive binding assay was purchased from ThermoFisher Scientific.
The construction of the pCI-puro vector and pCAGGS-gp350/220-F has been described (Ogembo et al., 2015; Perez et al., 2017).
Virus production and purification
eGFP-tagged EBV was produced from the EBV-infected AGS cell line as described (Ogembo et al., 2015). Briefly, AGS-EBV-eGFP cells were seeded to 90% confluency in T-75 flasks in Ham’s F-12 medium containing G418 antibiotic. After the cells reached confluency, G418 media was replaced with Ham’s F-12 medium containing 33 ng/ml 12-O-tetradecanoylphorbol-13-acetate (TPA) and 3 mM sodium butyrate (NaB) to induce lytic replication of the virus. Twenty-four h post-induction, the media was replaced with complete Ham’s F-12 media without G418, TPA, or NaB and cells were incubated for 4 days at 37°C in a 50% CO2 incubator. The cell supernatant was collected, centrifuged, and filtered using a 0.8-μm filter to remove cell debris. The filtered supernatant was ultra-centrifuged using a Beckman-Coulter type 19 rotor for 70 min at 14 941 xg to pellet the virus. EBV-eGFP virus was titrated in both HEK-293T cells and Raji cells, and stocks were stored at −80°C for subsequent experiments.
Generation and purification of gp350 virus-like particles (VLPs)
To generate gp350 VLPs, equal amounts (8 μg/plasmid) of the relevant plasmids (pCAGGS-Newcastle disease virus [NDV] matrix [M] and nucleocapsid proteins [NP], and gp350 ectodomain fused to NDV fusion [F] protein cytoplasmic and transmembrane domains) were co-transfected into 80% confluent CHO cells seeded in T-175cm2 flasks; supernatant from transfected cells containing VLPs was collected and VLPs were purified and composition characterized, as previously described (Ogembo et al., 2015).
Production of hybridoma cell lines
Seven days prior to immunization, two eight-week-old BALB/c mice were bled for collection of pre-immune serum. The mice were immunized with purified UV-inactivated EBV three times (Days 0, 21, and 35), then boosted with VLPs incorporating gp350 on the surface three times (Days 42, 49, and 56). The two mice were sacrificed and their splenocytes were isolated, purified, and fused with P3X63Ag8.653 myeloma cells at a ratio of 3:1 in the presence of polyethylene glycol (PEG, Sigma). Hybridoma cells were seeded in flat-bottom 96-well plates and selected in specialized hybridoma growth media with HAT (Sigma) and 10% FBS as described (Broering et al., 2009).
Indirect ELISA
Hybridoma cell culture supernatants from wells that had colony-forming cells were tested for antibody production using indirect ELISA. Briefly, immunoplates (Costar 3590; Corning Incorporated) were coated with 50 μl of 0.5 μg/ml recombinant EBV gp350 ectodomain (Immune Technology Corporation) diluted in 1x phosphate-buffered saline (PBS, pH 7.4) and incubated overnight at 4°C. After washing three times with 1x PBS containing 0.05% (v/v) Tween 20 (washing buffer), plates were blocked with 100 μl washing buffer containing 2% (w/v) bovine serum albumin (BSA) (blocking buffer), incubated for 1 h at room temperature, and washed as above. 100 μl of hybridoma supernatant or 50 μl of 10 μg/ml purified mAbs was added to each well (in triplicate) and incubated for 2 h at room temperature. Anti-KSHV gH/gL 54A1 and m72A1 mAbs were added as negative and positive controls, respectively. The plates were washed as described above, followed by incubation with 50 μl of goat anti-mouse IgG HRP-conjugated secondary antibody (1:2,000 diluted in 1x PBS) at room temperature for 1 h. The plates were washed again and 100 μl of chromogenic substrate 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulphonic acid) (ABTS, Life Science Technologies) was added. The reaction was stopped using 100 μl of ABTS peroxidase stop solution containing 5% sodium dodecyl sulfate (SDS) in water. The absorbance was read at an optical density of 405 nm using an ELISA reader (Molecular Devices). All experiments were performed in triplicate and confirmed in three independent experiments.
Antibody purification, quantification, and isotyping
Hybridoma cells from selected individual positive clones and filtered were expanded stepwise from 96-well plates to T-75 flasks. At confluence in T-75 flasks, supernatant from individual clones was collected, clarified by centrifugation (4,000 g, 10 min, 4°C), through a 0.22-μm membrane filter (Millipore). Antibodies were further purified using Capturem™ Protein A Maxiprep spin columns and stored in 1x PBS (pH 7.4) at 4°C. Alternatively, antibodies were purified using protein G affinity chromatography followed by size-exclusion chromatography at the Beckman Institute of City of Hope X-ray Crystallography Core facility. Antibodies were analyzed using SDS-PAGE to determine purity. Bicinchoninic acid assay (BCA assay; Thermo Fisher Scientific) was conducted to determine the concentration of purified antibodies. Isotype identification was performed using a Rapid ELISA mouse mAb isotyping kit (Thermo Fisher Scientific). Two independent experiments were performed.
RNA extraction, cDNA synthesis, and sequencing and analysis of the variable region of the mAbs
Total RNA was extracted from 1×106 hybridoma cells using the RNeasy Mini Kit (Qiagen). Each hybridoma clone cDNA was synthesized in a total volume of 20 μl using Tetro Reverse Transcriptase (200 u), RiboSafe RNase Inhibitor, Oligo(dT)18 primer, dNTP mix (10 mM each nucleotide), and 100–200 ng RNA. Reverse transcription was performed at 45°C for 30 min, and terminated at 85°C for 5 min. The cDNA was stored at −20°C. Immunoglobulin (Ig) VH and VL were amplified using the mouse Ig-specific primer set purchased from Novagen (Jones and Bendig, 1991). The VH and VL genes were amplified in separate reactions and PCR products were visualized on 1% agarose gels.
The VH and VL amplicons were sequenced using an Illumina MiSeq platform: duplicate 50-μl PCR reactions were performed, each containing 50 ng purified cDNA, 0.2 mM dNTPs, 1.5 mM MgCl2, 1.25 U Platinum Taq DNA polymerase, 2.5 μl 10x PCR buffer, and 0.5 μM of each primer designed to amplify the VH and VL. The amplicons were purified using an AxyPrep Mag PCR Clean-up kit (ThermoFisher Scientific). The Illumina primer PCR PE1.0 and index primers were used to allow multiplexing of samples. The library was quantified using ViiA™ 7 Real-Time PCR System (Life Technologies) and visualized for size validation on an Agilent 2100 Bioanalyzer (Agilent Technologies) using a high-sensitivity cDNA assay. The sequencing library pool was diluted to 4 nM and run on a MiSeq desktop sequencer (Illumina). The 600-cycle MiSeq Reagent Kit (Illumina) was used to run the 6 pM library with 20% PhiX (Illumina), and FASTQ files were used for data analysis (Pei et al., 2008). The determination of immunoglobulin families was analyzed using the IMGT/V-QUEST tool (www.imgt.org/IMGT_vquest/vquest) (Brochet et al., 2008).
Humanization of 72A1
To generate humanized mAbs, the BioLuminate interface (Schrödinger) was used to identify the human VH and VL framework using 72A1. The resulting model was visually inspected to ensure appropriate packing at the base of the CDR. The sequence was meditope-enabled to add functionality for generating bispecific antibodies in the future (Donaldson et al., 2013). The resulting sequences were codon-optimized, synthesized, and cloned into the PD2610 vector (Atum). The constructs were transiently transfected into ExpiCHO cells following the manufacturer’s protocol. Supernatant was collected at 10 days post-transfection and IgG was purified using protein G affinity chromatography, followed by size-exclusion chromatography.
Immunoblot analysis
CHO cells were cultured and stably co-transfected with full-length pCAGGS-gp350/220 and pCI-puro vector containing a puromycin resistance gene. Forty-eight h post-transfection, DMEM media containing 10 μg/ml of puromycin was added to enrich for cells expressing gp350/220 protein. Puromycin-resistant clones were expanded, followed by flow cytometry sorting using m72A1 to a purity >90%. EBV gp350-positive CHO cells were harvested and lysed in radioimmunoprecipitation assay buffer (RIPA) followed by centrifugation at 15,000 g for 15 min on a benchtop centrifuge. The lysate was collected and heated at 95°C for 10 min in SDS sample buffer containing β-mercaptoethanol, then separated using SDS-PAGE. Proteins were transferred onto a nitrocellulose membrane using an iBlot™ Transfer System (Thermo Fisher Scientific) followed by incubation in blocking buffer (1% BSA; 20 mM Tris–HCl, pH 7.5; 137 mM NaCl; and 0.1%Tween-20 [TBST]) for 1 h. The blots were incubated in TBST containing purified anti-gp350 antibodies (1:50) overnight at 4°C. After three washes with TBST, the blots were incubated with HRP-conjugated goat anti-mouse (1:2,000) in TBST for 1 h. After three washes, the antibody-protein complexes were detected using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare). All experiments were independently repeated three times.
Flow cytometry
To assess the ability of purified anti gp350 mAbs to detect surface expression of EBV gp350 protein by flow cytometry, CHO cells that stably express EBV gp350 were harvested and stained with purified anti-gp350 (10 μg/ml), followed by AF488 goat anti-mouse IgG secondary antibody. Flow cytometric analysis was performed on a C-6 FC (BD Biosciences) and data was analyzed using FlowJo Cytometry Analysis software (FlowJo, LLC) as described (Ogembo et al., 2015). All experiments were performed in triplicate and independently repeated three times.
EBV neutralization assay
EBV neutralization assay was performed in Raji cells as previously described (Ogembo et al., 2015). Briefly, purified individual anti-gp350 mAbs were incubated with purified AGS-EBV-eGFP (titer calculated to infect at least 8% of HEK-293 cells seeded in 100 μl of serum-free DMEM) for 2 h at 37°C. To represent EBV infection of B cells, the pre-incubated anti-gp350 mAbs/AGS-EBV-eGFP were used to infect 5×105 Raji cells seeded in a 96-well plate for 2 h at 37°C. Neutralizing 72A1 and non-neutralizing 2L10 anti-gp350 mAbs served as positive and negative controls, respectively. Infected cells were washed three times with PBS to remove any unbound viruses and antibodies. Washed, infected cells were incubated in 96-well plates at 37°C for 48 h post-infection and the number of eGFP+ (infected) cells was determined using flow cytometry. All dilutions were performed in quintuplicate and the assays repeated three times. Antibody EBV neutralization activity was calculated as: % neutralization = (EBValone - EBVmAb) / (EBValone) × 100.
Epitope mapping by competitive cell binding assay
To evaluate conformation epitope mapping of the selected mAbs, competitive binding assays were conducted using biotinylated mAbs. A one-step antibody biotinylation kit (MACS Miltenyi Biotec) was used to biotinylate the mAbs. Approximately 1 ×105 CHO cells that stably express EBV gp350 were incubated for 2 h with serially diluted (500, 250, 125, and 67.5 μg/ml) unlabeled competitor mAbs and non-specific anti-KSHV gH/gL 54A1 mAb. After being washed with PBS, the cells were incubated for 2 h in the presence of 1 μg/ml biotinylated mAbs. To determine maximum binding, cells in which the biotinylated mAb was added in the absence of unlabeled mAbs were included in the assay. Cells were washed with PBS, followed by incubation for 1 h with anti-biotin AF488 at a dilution of 1:500. After the final wash in PBS, cells were resuspended in 1% paraformaldehyde and analyzed by flow cytometry as described above. Percentage of inhibition was calculated as: 100 − [(% fluorescent cells with competitor mAb/% fluorescent cells without competitor mAb) × 100]. The complete prevention of binding of a biotinylated mAb by its unlabeled counterpart was used as a validation of the assay, as previously described (Chiuppesi et al., 2015).
Synthesis of 20-mer linear peptides of gp350 proteins
Forty-five sequential 20-mer linear peptides, covering the whole sequence of gp350 (GenBank: NC_007605.1), with an exception of aa 862–881, were synthesized using a solid phase method and cleaved using a low-high hydrogen fluoride method by the GenicBio, as previously described (Urquiza et al., 2005). Synthesis of aa 862–881 (pep-44) was not possible due to multiple hydrophobic aa.
Linear epitope mapping by peptide-mAb binding assay
The binding of anti-gp350 mAbs to 45 synthesized 20-mer sequential peptides covering the total length of gp350 was analyzed using indirect ELISA as described (Urquiza et al., 2005). Briefly, immunoplates were coated with 50 μl of 10 μg/ml EBV gp350 peptides (forty-five 20-mers) diluted in PBS and incubated overnight at 4°C; 0.5 μg/ml recombinant EBV gp350 ectodomain protein was used as a positive control. After washing three times with washing buffer (PBS containing 0.05% (v/v) Tween 20), plates were blocked with 100 μl washing buffer containing 3% BSA (blocking buffer), incubated for 1 h at room temperature, and washed as above. Ten μg/ml purified mAbs were added to each well in triplicate and incubated for 2 h at room temperature. Anti-gp350 antibodies m72A1 and h72A1 were added as positive controls and anti-KSHV-gH/gL 54A1 mAb was used as negative control. The plates were washed as described above, followed by incubation with goat anti-mouse IgG or goat anti-human IgG HRP-conjugated secondary antibody (1:2,000 diluted in PBS) at room temperature for 1 h. The plates were washed again and the chromogenic substrate ABTS was added. The reaction was stopped using ABTS peroxidase stop solution. The absorbance was read at an optical density of 405 nm using an ELISA plate reader.
Statistical Analysis
Unpaired Mann–Whitney U test was used to assess statistical differences between two independent groups. Statistical calculations were performed in Graphpad Prism. Data was considered statistically significant at p<0.05.
Supplementary Material
Highlights.
Biochemically and functionally characterized 15 new anti-gp350 murine mAbs
Identified four novel potent nAbs comparable to commercial 72A1 and 11 non-nAbs
The novel anti-gp350 nAbs bind similar overlapping epitopes (142–161) with 72A1
Humanized commercial nAb 72A1 to avoid human anti-murine antibody immune responses
The novel nAbs and humanized 72A1 have clinical potential to prevent EBV infection
Acknowledgments
We are grateful to Liisa Selin for providing EBV Akata virus expressing eGFP, and Dr. Yue Li of the City of Hope X-ray Crystallography Core facility for purification of antibodies using protein G affinity and size-exclusion chromatography. This work was supported by the National Institutes of Health (NIH) grant R21 CA232275, Toni Stephenson Lymphoma Center Pilot Grant award, and a generous philanthropic grant from the V Foundation to J.G.O. Research reported in this publication also included work performed at the Beckman Research Institute of City of Hope Integrative Genomics Core, Flow Cytometry Core, X-ray Crystallography Core and Animal Resource Center supported by the National Cancer Institute of the NIH under award number P30 CA033572. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. We thank Dr. Sarah T. Wilkinson for editing of the manuscript and for her insightful feedback and discussion.
Footnotes
Competing Interests: J.G.O and L.Z.M disclose that they have filed provisional patent, 62/491,945 related to this work. The other authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander BT, Hladnik LM, Augustin KM, Casabar E, McKinnon PS, Reichley RM, Ritchie DJ, Westervelt P, Dubberke ER, 2010. Use of Cytomegalovirus Intravenous Immune Globulin for the Adjunctive Treatment of Cytomegalovirus in Hematopoietic Stem Cell Transplant Patients. Pharmacotherapy 30, 554–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfarano C, Andrade CE, Anthony K, Bahroos N, Bajec M, Bantoft K, Betel D, Bobechko B, Boutilier K, Burgess E, Buzadzija K, Cavero R, D’Abreo C, Donaldson I, Dorairajoo D, Dumontier MJ, Dumontier MR, Earles V, Farrall R, Feldman H, Garderman E, Gong Y, Gonzaga R, Grytsan V, Gryz E, Gu V, Haldorsen E, Halupa A, Haw R, Hrvojic A, Hurrell L, Isserlin R, Jack F, Juma F, Khan A, Kon T, Konopinsky S, Le V, Lee E, Ling S, Magidin M, Moniakis J, Montojo J, Moore S, Muskat B, Ng I, Paraiso JP, Parker B, Pintilie G, Pirone R, Salama JJ, Sgro S, Shan T, Shu Y, Siew J, Skinner D, Snyder K, Stasiuk R, Strumpf D, Tuekam B, Tao S, Wang Z, White M, Willis R, Wolting C, Wong S, Wrong A, Xin C, Yao R, Yates B, Zhang S, Zheng K, Pawson T, Ouellette BF, Hogue CW, 2005. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic acids research 33, D418–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA, 1998. EBV persistence in memory B cells in vivo. Immunity 9, 395–404. [DOI] [PubMed] [Google Scholar]
- Baer R, Bankier AT, Biggin MD, Deininger PL, Farrell PJ, Gibson TJ, Hatfull G, Hudson GS, Satchwell SC, Seguin C, Tuffnell PS, Barrell BG, 1984. DNA-Sequence and Expression of the B95–8 Epstein-Barr Virus Genome. Nature 310, 207–211. [DOI] [PubMed] [Google Scholar]
- Benkerrou M, Jais JP, Leblond V, Durandy A, Sutton L, Bordigoni P, Garnier JL, Le Bidois J, Le Deist F, Blanche S, Fischer A, 1998. Anti-B-cell monoclonal antibody treatment of severe posttransplant B-lymphoproliferative disorder: prognostic factors and long-term outcome. Blood 92, 3137–3147. [PubMed] [Google Scholar]
- Brochet X, Lefranc MP, Giudicelli V, 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res 36, W503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broering TJ, Garrity KA, Boatright NK, Sloan SE, Sandor F, Thomas WD Jr., Szabo G, Finberg RW, Ambrosino DM, Babcock GJ, 2009. Identification and characterization of broadly neutralizing human monoclonal antibodies directed against the E2 envelope glycoprotein of hepatitis C virus. J Virol 83, 12473–12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W, Joyce MG, Nguyen H, Banh DV, Aguilar F, Tariq Z, Yap ML, Tsujimura Y, Gillespie RA, Tsybovsky Y, Andrews SF, Narpala SR, McDermott AB, Rossmann MG, Yasutomi Y, Nabel GJ, Kanekiyo M, Cohen JI, 2019. Immunization with Components of the Viral Fusion Apparatus Elicits Antibodies That Neutralize Epstein-Barr Virus in B Cells and Epithelial Cells. Immunity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sathiyamoorthy K, Zhang X, Schaller S, Perez White BE, Jardetzky TS, Longnecker R, 2018. Ephrin receptor A2 is a functional entry receptor for Epstein-Barr virus. Nat Microbiol 3, 172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesnokova LS, Nishimura SL, Hutt-Fletcher LM, 2009. Fusion of epithelial cells by Epstein–Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins αvβ6 or αvβ8. Proceedings of the National Academy of Sciences 106, 20464–20469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiuppesi F, Wussow F, Johnson E, Bian C, Zhuo M, Rajakumar A, Barry PA, Britt WJ, Chakraborty R, Diamond DJ, 2015. Vaccine-Derived Neutralizing Antibodies to the Human Cytomegalovirus gH/gL Pentamer Potently Block Primary Cytotrophoblast Infection. J Virol 89, 11884–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JI, 2000. Epstein-Barr virus infection. New England Journal of Medicine 343, 481–492. [DOI] [PubMed] [Google Scholar]
- Cohen JI, 2015. Epstein-Barr virus vaccines. Clin Transl Immunology 4, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JI, 2018. Vaccine Development for Epstein-Barr Virus. Adv Exp Med Biol 1045, 477–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JI, Fauci AS, Varmus H, Nabel GJ, 2011. Epstein-Barr Virus: An Important Vaccine Target for Cancer Prevention. Science Translational Medicine 3, 107fs107–107fs107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JI, Mocarski ES, Raab-Traub N, Corey L, Nabel GJ, 2013. The need and challenges for development of an Epstein-Barr virus vaccine. Vaccine 31 Suppl 2, B194–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Jackson JO, Jardetzky TS, Longnecker R, 2011. Fusing structure and function: a structural view of the herpesvirus entry machinery: A structural view of herpesvirus entry machinery. Nat Rev Microbiol 9, 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coté TR, Biggar RJ, Rosenberg PS, Devesa SS, Percy C, Yellin FJ, Lemp G, Hardy C, Geodert JJ, Blattner WA, 1998. Non‐Hodgkin’s lymphoma among people with AIDS: Incidence, presentation and public health burden. International Journal of Cancer 73, 645–650. [DOI] [PubMed] [Google Scholar]
- Cruz RJ Jr., Ramachandra S, Sasatomi E, DiMartini A, de Vera M, Fontes P, Hughes C, Humar A, 2012. Surgical management of gastrointestinal posttransplant lymphoproliferative disorders in liver transplant recipients. Transplantation 94, 417–423. [DOI] [PubMed] [Google Scholar]
- Cui X, Cao Z, Sen G, Chattopadhyay G, Fuller DH, Fuller JT, Snapper DM, Snow AL, Mond JJ, Snapper CM, 2013. A novel tetrameric gp350 1–470 as a potential Epstein-Barr virus vaccine. Vaccine 31, 3039–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JM, Zer C, Avery KN, Bzymek KP, Horne DA, Williams JC, 2013. Identification and grafting of a unique peptide-binding site in the Fab framework of monoclonal antibodies. Proc Natl Acad Sci U S A 110, 17456–17461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH, 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4, 800–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels EA, Pfeiffer RM, Fraumeni JF Jr., Kasiske BL, Israni AK, Snyder JJ, Wolfe RA, Goodrich NP, Bayakly AR, Clarke CA, Copeland G, Finch JL, Fleissner ML, Goodman MT, Kahn A, Koch L, Lynch CF, Madeleine MM, Pawlish K, Rao C, Williams MA, Castenson D, Curry M, Parsons R, Fant G, Lin M, 2011. Spectrum of cancer risk among US solid organ transplant recipients. JAMA 306, 1891–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faro A, Kurland G, Michaels MG, Dickman PS, Greally PG, Spichty KJ, Noyes BB, Boas SR, Fricker FJ, Armitage JM, Zeevi A, 1996. Interferon-alpha affects the immune response in post-transplant lymphoproliferative disorder. Am J Respir Crit Care Med 153, 1442–1447. [DOI] [PubMed] [Google Scholar]
- Fingeroth JD, Weis JJ, Tedder TF, Strominger JL, Biro PA, Fearon DT, 1984. Epstein-Barr virus receptor of human B lymphocytes is the C3d receptor CR2. Proc Natl Acad Sci U S A 81, 4510–4514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotz D, Chapman JR, Dharnidharka VR, Hanto DW, Castro MC, Hirsch HH, Leblond V, Mehta AK, Moulin B, Pagliuca A, 2012. The seville expert workshop for progress in posttransplant lymphoproliferative disorders. Transplantation 94, 784–793. [DOI] [PubMed] [Google Scholar]
- Goedert JJ, Cote TR, Virgo P, Scoppa SM, Kingma DW, Gail MH, Jaffe ES, Biggar RJ, 1998. Spectrum of AIDS-associated malignant disorders. Lancet 351, 1833–1839. [DOI] [PubMed] [Google Scholar]
- Gu SY, Huang TM, Ruan L, Miao YH, Lu H, Chu CM, Motz M, Wolf H, 1995. First EBV vaccine trial in humans using recombinant vaccinia virus expressing the major membrane antigen. Dev Biol Stand 84, 171–177. [PubMed] [Google Scholar]
- Han T, Marasco WA, 2011. Structural basis of influenza virus neutralization. Annals of the New York Academy of Sciences 1217, 178–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque T, Johannessen I, Dombagoda D, Sengupta C, Burns DM, Bird P, Hale G, Mieli-Vergani G, Crawford DH, 2006. A mouse monoclonal antibody against Epstein-Barr virus envelope glycoprotein 350 prevents infection both in vitro and in vivo. J Infect Dis 194, 584–587. [DOI] [PubMed] [Google Scholar]
- Henle G, Henle W, Diehl V, 1968. Relation of Burkitt’s tumor-associated herpes-ytpe virus to infectious mononucleosis. Proceedings of the National Academy of Sciences of the United States of America 59, 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrman M, Muhe J, Quink C, Wang F, 2015. Epstein-Barr Virus gp350 Can Functionally Replace the Rhesus Lymphocryptovirus Major Membrane Glycoprotein and Does Not Restrict Infection of Rhesus Macaques. J Virol 90, 1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrman M, Muhe J, Quink C, Wang F, 2016. Epstein-Barr Virus gp350 Can Functionally Replace the Rhesus Lymphocryptovirus Major Membrane Glycoprotein and Does Not Restrict Infection of Rhesus Macaques. J Virol 90, 1222–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GJ, Lazarowitz SG, Hayward SD, 1980. Monoclonal antibody against a 250,000-dalton glycoprotein of Epstein-Barr virus identifies a membrane antigen and a neutralizing antigen. Proc Natl Acad Sci U S A 77, 2979–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, 2016. Exploiting Mucosal Immunity for Antiviral Vaccines. Annu Rev Immunol 34, 575–608. [DOI] [PubMed] [Google Scholar]
- Jones ST, Bendig MM, 1991. Rapid PCR-cloning of full-length mouse immunoglobulin variable regions. Biotechnology (N Y) 9, 579. [DOI] [PubMed] [Google Scholar]
- Kanekiyo M, Bu W, Joyce MG, Meng G, Whittle JR, Baxa U, Yamamoto T, Narpala S, Todd JP, Rao SS, McDermott AB, Koup RA, Rossmann MG, Mascola JR, Graham BS, Cohen JI, Nabel GJ, 2015. Rational Design of an Epstein-Barr Virus Vaccine Targeting the Receptor-Binding Site. Cell 162, 1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Sherritt M, Burrows SR, 1999. EBV structural antigens, gp350 and gp85, as targets for ex vivo virus-specific CTL during acute infectious mononucleosis: potential use of gp350/gp85 CTL epitopes for vaccine design. The Journal of Immunology 162, 3063–3069. [PubMed] [Google Scholar]
- Ladner RC, 2007. Mapping the epitopes of antibodies. Biotechnol Genet Eng Rev 24, 1–30. [DOI] [PubMed] [Google Scholar]
- Milpied N, Vasseur B, Parquet N, Garnier JL, Antoine C, Quartier P, Carret AS, Bouscary D, Faye A, Bourbigot B, Reguerre Y, Stoppa AM, Bourquard P, Hurault de Ligny B, Dubief F, Mathieu-Boue A, Leblond V, 2000. Humanized anti-CD20 monoclonal antibody (Rituximab) in post transplant B-lymphoproliferative disorder: a retrospective analysis on 32 patients. Ann Oncol 11 Suppl 1, 113–116. [PubMed] [Google Scholar]
- Moutschen M, Leonard P, Sokal EM, Smets F, Haumont M, Mazzu P, Bollen A, Denamur F, Peeters P, Dubin G, Denis M, 2007. Phase I/II studies to evaluate safety and immunogenicity of a recombinant gp350 Epstein-Barr virus vaccine in healthy adults. Vaccine 25, 4697–4705. [DOI] [PubMed] [Google Scholar]
- Mulama DH, Mutsvunguma LZ, Totonchy J, Ye P, Foley J, Escalante GM, Rodriguez E, Nabiee R, Muniraju M, Wussow F, Barasa AK, G. OJ, 2019. A multivalent Kaposi sarcoma-associated herpesvirus-like particle vaccine capable of eliciting high titers of neutralizing antibodies in immunized rabbits. Vaccine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemerow GR, Houghten RA, Moore MD, Cooper NR, 1989. Identification of an epitope in the major envelope protein of Epstein-Barr virus that mediates viral binding to the B lymphocyte EBV receptor (CR2). Cell 56, 369–377. [DOI] [PubMed] [Google Scholar]
- Nemerow GR, Mold C, Schwend VK, Tollefson V, Cooper NR, 1987a. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J Virol 61, 1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemerow GR, Mold C, Schwend VK, Tollefson V, Cooper NR, 1987b. Identification of Gp350 as the Viral Glycoprotein Mediating Attachment of Epstein-Barr-Virus (Ebv) to the Ebv/C3d Receptor of B-Cells - Sequence Homology of Gp350 and C3-Complement Fragment C3d. Journal of Virology 61, 1416–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogembo JG, Kannan L, Ghiran I, Nicholson-Weller A, Finberg RW, Tsokos GC, Fingeroth JD, 2013. Human complement receptor type 1/CD35 is an Epstein-Barr Virus receptor. Cell Rep 3, 371–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogembo JG, Muraswki MR, McGinnes LW, Parcharidou A, Sutiwisesak R, Tison T, Avendano J, Agnani D, Finberg RW, Morrison TG, Fingeroth JD, 2015. A chimeric EBV gp350/220-based VLP replicates the virion B-cell attachment mechanism and elicits long-lasting neutralizing antibodies in mice. J Transl Med 13, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos EB, Ladanyi M, Emanuel D, Mackinnon S, Boulad F, Carabasi MH, Castro-Malaspina H, Childs BH, Gillio AP, Small TN, et al. , 1994. Infusions of donor leukocytes to treat Epstein-Barr virus-associated lymphoproliferative disorders after allogeneic bone marrow transplantation. N Engl J Med 330, 1185–1191. [DOI] [PubMed] [Google Scholar]
- Pei J, Kim B-H, Grishin NV, 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Research 36, 2295–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EM, Foley J, Tison T, Silva R, Ogembo JG, 2016. Novel Epstein-Barr virus-like particles incorporating gH/gL-EBNA1 or gB-LMP2 induce high neutralizing antibody titers and EBV-specific T-cell responses in immunized mice. Oncotarget. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez EM, Foley J, Tison T, Silva R, Ogembo JG, 2017. Novel Epstein-Barr virus-like particles incorporating gH/gL-EBNA1 or gB-LMP2 induce high neutralizing antibody titers and EBV-specific T-cell responses in immunized mice. Oncotarget 8, 19255–19273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piedimonte G, King KA, Holmgren NL, Bertrand PJ, Rodriguez MM, Hirsch RL, 2000. A humanized monoclonal antibody against respiratory syncytial virus (palivizumab) inhibits RSV-induced neurogenic-mediated inflammation in rat airways. Pediatr Res 47, 351–356. [DOI] [PubMed] [Google Scholar]
- Qualtiere LF, Decoteau JF, Hassan Nasr-el-Din M, 1987a. Epitope mapping of the major Epstein-Barr virus outer envelope glycoprotein gp350/220. J Gen Virol 68 ( Pt 2), 535–543. [DOI] [PubMed] [Google Scholar]
- Qualtiere LF, Decoteau JF, Nasreldin MH, 1987b. Epitope Mapping of the Major Epstein-Barr-Virus Outer Envelope Glycoprotein Gp350/220. Journal of General Virology 68, 535–543. [DOI] [PubMed] [Google Scholar]
- Rees L, Tizard EJ, Morgan AJ, Cubitt WD, Finerty S, Oyewole-Eletu TA, Owen K, Royed C, Stevens SJ, Shroff RC, Tanday MK, Wilson AD, Middeldorp JM, Amlot PL, Steven NM, 2009. A phase I trial of epstein-barr virus gp350 vaccine for children with chronic kidney disease awaiting transplantation. Transplantation 88, 1025–1029. [DOI] [PubMed] [Google Scholar]
- Rickinson AB, Kieff E, 2007a. Epstein-Barr Virus, in: Knipe DM, Howley PM (Eds.), Fields Virology. Lippiincott, Williams & Wilkins, Philadelphia, pp. 2655–2700. [Google Scholar]
- Rickinson AB, Kieff E, 2007b. Epstein-Barr Virus, in: Knipe D, Howley P (Eds.), Fields Virology, Fifth ed. Lippincott Wilkins and Williams, Philadelphia, pp. 2680–2700. [Google Scholar]
- Sashihara J, Burbelo PD, Savoldo B, Pierson TC, Cohen JI, 2009. Human antibody titers to Epstein-Barr Virus (EBV) gp350 correlate with neutralization of infectivity better than antibody titers to EBV gp42 using a rapid flow cytometry-based EBV neutralization assay. Virology 391, 249–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela M, Schechter B, Schechter I, Borek F, 1967. Antibodies to Sequential and Conformational Determinants. Cold Spring Harbor Symposia on Quantitative Biology 32, 537–545. [Google Scholar]
- Sitompul LS, Widodo N, Djati MS, Utomo DH, 2012. Epitope mapping of gp350/220 conserved domain of epstein barr virus to develop nasopharyngeal carcinoma (npc) vaccine. Bioinformation 8, 479–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snijder J, Ortego MS, Weidle C, Stuart AB, Gray MD, McElrath MJ, Pancera M, Veesler D, McGuire AT, 2018. An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus. Immunity 48, 799–811 e799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal EM, Hoppenbrouwers K, Vandermeulen C, Moutschen M, Leonard P, Moreels A, Haumont M, Bollen A, Smets F, Denis M, 2007. Recombinant gp350 vaccine for infectious mononucleosis: a phase 2, randomized, double-blind, placebo-controlled trial to evaluate the safety, immunogenicity, and efficacy of an Epstein-Barr virus vaccine in healthy young adults. J Infect Dis 196, 1749–1753. [DOI] [PubMed] [Google Scholar]
- Stamatatos L, Morris L, Burton DR, Mascola JR, 2009. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nature medicine 15, 866–870. [DOI] [PubMed] [Google Scholar]
- Starzl TE, Holmes JH, 1964. Experience in renal transplantation.
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen L. m., Santelli E, Stec B, Cadwell G, Ali M, 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nature structural & molecular biology 16, 265–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi G, Klein MG, Hannan JP, Young KA, Ma RZ, Asokan R, Holers VM, Chen XS, 2006. Structure of the Epstein-Barr virus major envelope glycoprotein. Nat Struct Mol Biol 13, 996–1001. [DOI] [PubMed] [Google Scholar]
- Tanner J, Weis J, Fearon D, Whang Y, Kieff E, 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50, 203–213. [DOI] [PubMed] [Google Scholar]
- Tanner J, Whang Y, Sample J, Sears A, Kieff E, 1988. Soluble gp350/220 and deletion mutant glycoproteins block Epstein-Barr virus adsorption to lymphocytes. J Virol 62, 4452–4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JE, Coincon M, Leblond V, Hu J, Fang JM, Sygusch J, Alfieri C, 2015. Peptides designed to spatially depict the Epstein-Barr virus major virion glycoprotein gp350 neutralization epitope elicit antibodies that block virus-neutralizing antibody 72A1 interaction with the native gp350 molecule. J Virol 89, 4932–4941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JE, Hu J, Alfieri C, 2018. Construction and Characterization of a Humanized Anti-Epstein-Barr Virus gp350 Antibody with Neutralizing Activity in Cell Culture. Cancers (Basel) 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA, Geilinger K, 1980. Monoclonal antibodies against the major glycoprotein (gp350/220) of Epstein-Barr virus neutralize infectivity. Proc Natl Acad Sci U S A 77, 5307–5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorley-Lawson DA, Poodry CA, 1982. Identification and isolation of the main component (gp350-gp220) of Epstein-Barr virus responsible for generating neutralizing antibodies in vivo. J Virol 43, 730–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugizov SM, Berline JW, Palefsky JM, 2003. Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat Med 9, 307–314. [DOI] [PubMed] [Google Scholar]
- Urquiza M, Lopez R, Patino H, Rosas JE, Patarroyo ME, 2005. Identification of three gp350/220 regions involved in Epstein-Barr virus invasion of host cells. J Biol Chem 280, 35598–35605. [DOI] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss ER, Alter G, Ogembo JG, Henderson JL, Tabak B, Bakis Y, Somasundaran M, Garber M, Selin L, Luzuriaga K, 2017. High Epstein-Barr Virus Load and Genomic Diversity Are Associated with Generation of gp350-Specific Neutralizing Antibodies following Acute Infectious Mononucleosis. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng N-Y, Mays I, Garman L, Helms C, 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453, 667–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanik EL, Smith JM, Shiels MS, Clarke CA, Lynch CF, Kahn AR, Koch L, Pawlish KS, Engels EA, 2017. Cancer Risk After Pediatric Solid Organ Transplantation. Pediatrics 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Li Y, Wang HB, Zhang A, Chen ML, Fang ZX, Dong XD, Li SB, Du Y, Xiong D, He JY, Li MZ, Liu YM, Zhou AJ, Zhong Q, Zeng YX, Kieff E, Zhang Z, Gewurz BE, Zhao B, Zeng MS, 2018. Ephrin receptor A2 is an epithelial cell receptor for Epstein-Barr virus entry. Nat Microbiol 3, 1–8. [DOI] [PubMed] [Google Scholar]
- Zhang PF, Klutch M, Armstrong G, Qualtiere L, Pearson G, Marcus-Sekura CJ, 1991. Mapping of the epitopes of Epstein-Barr virus gp350 using monoclonal antibodies and recombinant proteins expressed in Escherichia coli defines three antigenic determinants. J Gen Virol 72 ( Pt 11), 2747–2755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.