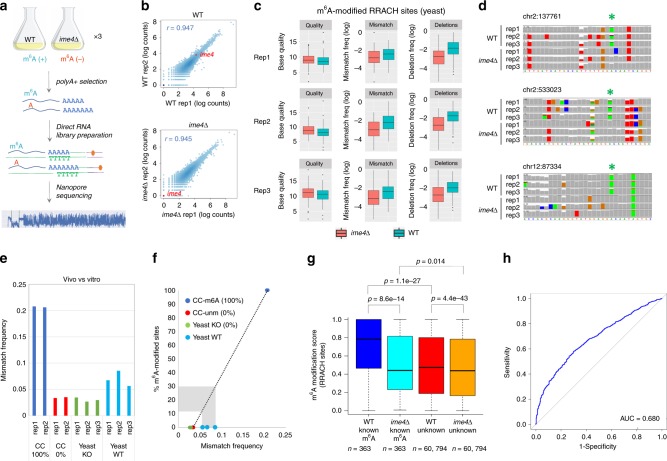
Fig. 3.
Yeast wild-type and ime4∆ strains show distinct base-called features at known m6A-modified RRACH sites. a Overview of the direct RNA sequencing library preparation using in vivo polyA(+) RNA from S. cerevisiae cultures. b Replicability of per-gene counts using direct RNA sequencing across wild-type yeast strains (top) and ime4∆ strains (middle). The correlation between wild-type and ime4∆ strains is also shown (bottom). c Comparison of the observed mismatch frequencies in the 100%-modified in vitro transcribed sequences (blue), unmodified sequences (red), yeast ime4∆ knockout (green), and yeast wild type (cyan). Values for each biological replicate are shown. d Base-called features (base quality, insertion frequency, and deletion frequency) of RRACH 5-mers known to contain m6A modifications. Only features corresponding to the modified nucleotide (position 0) are shown. Features extracted from wild-type yeast reads (m6A-modified) are shown in blue, whereas those from ime4∆ (unmodified) for the same set of k-mers are shown in red. f Genomic tracks of previously reported m6A-modified RRACH sites in yeast, identified using Illumina sequencing. The m6A-modified nucleotide is highlighted with a green asterisk. In these positions, wild-type yeast strains show increased mismatch frequencies, as well as decreased coverage — reflecting increased deletion frequency — in all three biological replicates, whereas these features are not observed in any of the three ime4∆ replicates. g Predicted m6A modification scores predicted by the trained SVM at known m6A-modified (n = 363) and unknown (n = 60,794) RRACH sites, both for yeast wild-type and ime4∆ data sets. P-values have been computed using Kruskal–Wallis test. A site was included in the analysis if there were mapped reads present in all six yeast samples. Sites with more than one “A” in the 5-mer were excluded from the analysis. h ROC curve depicting the performance of EpiNano in yeast data sets (n = 61,363 sites). Error bars indicate s.d. Source data are provided in the Source Data file