Abstract
The steadily increasing epidemic of obesity continues at alarming rates, is an important public health problem, and expression changes of S100A16 and 11 β-hydroxysteroid dehydrogenase type 1(11β-HSD1) is attributable to the adipocyte differentiation. In our previous study, we found that 11β-HSD1 protein expression increased in S100A16-overexpressed 3T3-L1 cell model. In order to further investigate the relationship between S100A16 and 11β-HSD1, and the molecular mechanisms of S100A16-induced adipogenesis, we constructed S100A16 transgenic and knockout mouse, and S100A16-overexpressed 3T3-L1 preadipocyte cell. Using S100A16 transgenic (S100A16Tg/+) mice fed with normal fat diet (NFD) and high fat diet (HFD) diet model, we evaluated the effect of S100A16 on adipogenesis, expression of 11β-HSD1, and RNA sequencing and quantification of gene expression. Using the 3T3-L1 cell model, we examined the effect of S100A16 and 11β-HSD1 on pre-adipocyte differentiation, and cell signaling events of 11β-HSD1 overexpression induced by S100A16. We found that when compared with C57BL/6 mice, overexpression of S100A16 under the condition of HFD increased lipid content in WAT and fat infiltration in hepatocytes, 11β-HSD1 protein expression increased along with S100A16. Elevated S100A16 and 11β-HSD1 expression promoted adipogenesis in 3T3-L1 cells. Overexpression of S100A16 inhibited the degradation of 11β-HSD1. We conclude that S100A16-induced adipogenesis is associated with up-regulation of 11β-HSD1.
Keywords: 11β-HSD1, adipogenesis, obesity, S100A16, type 2 diabetes
Introduction
Obesity is an important metabolic disorder and serious public health problem worldwide that is closely associated with various dangerous disease risk factors, such as insulin resistance and Type 2 diabetes. Due to the wide-ranging health implications, the need to better understand the cellular and molecular basis of adipocyte differentiation in order to develop new and effective strategies for obesity prevention has become acute [1,2]. During adipocyte differentiation, chronological changes in the expression of numerous genes take place throughout the process, including C/EBP-α, PPAR-γ, S100A16, and 11β-HSD1 [3–5].
S100 proteins are small, acidic calcium-binding proteins of 10–12 kDa in size that contain two distinct EF-hand motifs. The S100-specific EF-hand is located at the N-terminus, followed by a classical Ca2+-binding EF-hand that operates as a Ca2+-activated switch that interacts with and modulates the activity of a large number of targets [6]. The S100 protein family is involved in the regulation of diverse cellular processes such as cell growth, differentiation, and cell cycle progression. The recently identified S100 protein family member S100A16 has been linked with obesity, Type 2 diabetes, inflammation, and cancer, via a Ca2+-dependent mechanism [4,7–12]. S100A16 is expressed widely in human tissues, and its precise biological functions are not fully understood.
The enzyme hydroxysteroid dehydrogenase type 1 (11β-HSD1) is a key enzyme that catalyzes the intracellular conversion of cortisone to physiologically active cortisol [13,14], and this enzyme has received widespread attention in the past decade. A series of studies showed that 11β-HSD1 can promote preadipocyte differentiation and adipogenesis, and cause insulin resistance [15,16]. Additionally, overexpression of 11β-HSD1 in adipose tissue is a key feature of metabolic syndrome [17,18].
In our present study, we demonstrated that up-regulation of S100A16 expression in adipose tissue promoted 11β-HSD1 expression, while down-regulation suppressed 11β-HSD1 expression, suggesting S100A16 may be associated with pre-adipocyte differentiation, obesity, and insulin resistance. We used an S100A16 transgenic (S100A16Tg/+) mouse model fed with a high fat diet (HFD) to investigate the effect of S100A16 on adipogenesis and the expression of 11β-HSD1. Cultured adipocytes were used to investigate the effect of S100A16 and 11β-HSD1 on preadipocyte differentiation, and the molecular mechanism of up-regulation of 11β-HSD1 induced by overexpression of S100A16 was explored.
Materials and methods
Mouse breeding and genotyping
S100A16 transgenic heterozygous mouse (S100A16Tg/+) and S100A16 knockout heterozygous mouse (S100A16KO/+) were generated at the Model Animal Research Center Of Nanjing University (Contract [2008] 054, [2009] T 67). The promoter used for S100A16Tg/+ mouse was CAG. The genetic background of S100A16Tg/+ and S100A16KO/+ was C57BL/6. All mice were housed, fed, and monitored at the Model Animal Research Center of Nanjing University (MARC), with 12-h light/dark cycles and with free access to food and water. The facility is maintained in a constant temperature and humidity environmental conditions. The Ethics Committees, the Institutional Animal Care and Use Committee (IACUC) at the University of Nanjing Medical University, approved all animal protocols. Genotyping was carried out using primers listed in Supplementary Figures S1 and S3B. Unfortunately, we were unable to obtain sufficient heterozygous S100A16KO/+ mice, presumably because knockdown of S100A16 may have affected their reproductive capacity. We cannot get homozygous S100A16KO/ KO mouse. And this is being investigated separately within our group.
A total of 10 C57BL/6 and 10 S100A16Tg/+ 5-week-old male mice were fed with a NFD or HFD diet for 17 weeks (from 5 to 21 weeks of age). The HFD was supplied by OpenSource DIETS (Research Diets, Inc., New Brunswick, NJ, U.S.A., #D12451, the details of the NFD is as Supplementary Table S1). We monitored the body weight of all mice every week and measured the visceral fat weight following administration of the anesthetic Nembutal (100 mg/kg). In addition, we investigated the morphology of visceral fat cells from mice fed normal chow and a HFD using hematoxylin and eosin staining.
Cell culture and differentiation
The 3T3-L1 mouse preadipocyte cell line was purchased from ATCC and cultured in high glucose Dulbecco’s modified Eagle medium (DMEM; Thermo Fisher) containing 10% newborn calf serum, 100 U/ml penicillin, and 100 mg/ml streptomycin in 5% CO2 at 37°C. Preadipocyte differentiation was induced as described previously [19]. To induce adipocyte differentiation, 3T3-L1 cells were seeded and allowed to grow for 2 days to reach confluence (designated as day 0), then cultured with DMEM containing 10% FBS and 0.5 mM 3-isobutyl-1-methyxanthine, 1 μg/ml porcine insulin, and 1 mM dexamethasone (MIX, Sigma). After 48 h of incubation (designated as day 2), the medium was replaced with DMEM containing 10% FBS and 1 μg/ml insulin. On day 4, cells were cultured with DMEM containing 10% FBS and the incubation was continued for 4 days with two changes of medium.
Construction of S100A16 and 11β-HSD1 interference expression plasmids, transfection, and stable clone selection
Four distinct domains within the coding region of mouse S100A16 (NM_026416.2) and 11β-HSD1 (NM_001044751) cDNAs were targeted for RNA interference. For this purpose, four pairs of reverse complementary oligonucleotides targeting mouse S100A16 were designed and sequences are given in Supplementary Table S2. Selected regions of 11β-HSD1 were also targeted by siRNA using the primers of Supplementary Table S3. Briefly, total RNA was purified using Trizol Reagent (Invitrogen).
Oligonucleotides were annealed and inserted into the pcDNA6.2-GW/EmGFP-miR expression vector (Invitrogen, #K4936-00). And then we generated pcDNA6.2-GW/EmGFP-miR-S100A16-1, 2, 3, and 4, and pcDNA6.2-GW/EmGFP-miR-11β-HSD1-1, 2, 3, and 4. A control construct was also created.
We used X-tremeGENE HP DNA Transfection Reagent (Roche, (#06365752001) to separately transfect the nine different plasmids into 3T3-L1 cells. To select for successful transfectants, cells were cultured at 24 h after transfection in selection medium containing 4 μg/ml blasticidin (Sigma). Blasticidin-resistant cells were maintained in culture medium supplemented with 2 μg/ml blasticidin for further analysis.
Construction of S100A16 and 11β-HSD1 overexpression plasmids, transfection and stable clone selection
Overexpression plasmid pcDNA3.1(+)-S100A16 and pcDNA3.1(+)-11β-HSD1 were constructed by synthesizing full-length mouse S100A16 (NM_026416.2) and 11β-HSD1 (NM_001044751) cDNAs and cloning into pcDNA3.1(+) using HindIII and EcoRI restriction sites. Plasmid DNA was purified from transformed bacteria and the final construct was verified by sequencing. The sequence congruence within the used restriction sites was 100%.
Expression constructs were transfected into 3T3-L1 preadipocytes using X-tremeGENE HP DNA Transfection Reagent (Roche, #06365752001). After 48 h, cells were cultured in selective DMEM medium containing 600 μg/ml G418 (Sigma) for 2 weeks for the selection of resistant colonies. G418-resistant cells were maintained in culture medium supplemented with 300 μg/ml G418 for further analysis.
Oil Red O staining
Oil Red O (O0625) was purchased from Sigma-Aldrich (St. Louis, MO). Differentiated 3T3-L1 cells (day 0, 4, or 10) were washed three times with PBS and stained with filtered Oil Red O solution (stock solution, 3 mg/ml in isopropanol; working solution, 60% stock solution and 40% distilled water) for 60 min at room temperature. Cells were then washed with ddH2O to remove unbound dye, and visualized and photographed under a microscope.
Protein extraction and Western blotting analysis
Tissues and cells were washed twice with ice-cold PBS, and 100 mg tissue was lysed with 1 ml lysis buffer (50 mM TRIS-HCl pH 7.4, 150 mM NaCl, 1% v/v Nonidet-P40, 1 mM EDTA, 1 mM NaF, 10 mg/ml aprotinin, 10 mM leupeptin, and 1 mM phenylmethanesulfonyl fluoride). Cells were scraped into lysis buffer, and tissues and cells were incubated on ice for 30 min. After centrifugation at 4°C, proteins in the supernatant were extracted, and the concentration was detected using BCA Protein Assay Kit (23225, PIERCE, U.S.A.). The protein was separated by SDS-PAGE then subjected to a standard Western blotting assay and imaged using a Molecular Imager ChemiDoc XRSC with the Image Lab Software (Version 4.0.1, Bio-Rad Laboratories, Hercules, CA, U.S.A.). Antibodies for 11β-HSD1 (AF3397) were purchased from R&D SYSTEMS, α-Tubulin (T5168) antibody was purchased from Sigma-Aldrich (St. Louis, MO), and S100A16 antibody (ab130419) was purchased from Abcam (U.S.A.).
Intraperitoneal glucose tolerance test (IGTT) and insulin tolerance test (ITT)
After overnight fasting, the tail blood glucose concentration (mM) was recorded (designated time = 0 min), mice were injected intraperitoneally with glucose at a dose of 2 g/kg of body weight, and blood glucose levels were monitored using a handheld glucometer (ACCU-CHEK Performa, Roche) at 15, 30, 60, and 120 min.
For ITTs, all mice were starved for 12 h, injected with a bolus of insulin (0.3 unit/kg of body weight), and their blood glucose levels were monitored using a handheld glucometer (ACCU-CHEK Performa, Roche) at 15, 30, 60, and 120 min.
RNA sequencing and quantification of gene expression
Adipose tissues from C57BL/6, S100A16KO/+ and S100A16Tg/+ (n = 3) mice were used for total RNA extraction using TRIzol reagent (Invitrogen), and RNA quality was tested using a Bioanalyzer 2200 (Agilent) instrument, samples with RIN ≥ 8.0 were used for RNA sequencing by BGI Tech Solutions. Briefly, total RNA was treated with Dnase I, mRNA was enriched using oligo(dT) magnetic beads. Then, the mRNA fragments is used for synthesis of the first and second strands of cDNA. The double-stranded cDNA products were sequenced using a Illumina HiSeq 2000. The RNA seq data analysis was also performed by BGI Tech Solutions.
Mouse embryonic fibroblasts (MEFs)
MEFs were isolated from Wild-type (C57BL/6), S100A16Tg/Tg and S100A16KO/+ mouse embryos at 13.5 days post coitum. Briefly, embryos were chopped into pieces and incubated in 0.025% trypsin and 0.5 mM EDTA at 37°C for 60 min with periodic agitation. Cells were washed with DMEM containing 10% FBS and dispersed by pipeting. S100A16 protein levels were determined using Western blotting. Cells were treated with 20 μM cycloheximide (CHX; C7698, Sigma) for 0, 12, or 24 h, and 11β-HSD1 protein expression was analyzed by Western blotting.
Triglyceride GPO-POD assay
Cellular triglyceride content was determined by using a Triglyceride GPO-POD Assay Kit (Sigma) according to a previously published method [4]. 3T3-L1 cells were cultured and induced in 10-cm well to differentiate into adipocytes (0 d, 4 d, and 10 d) before being washed with PBS twice, scraped in 500 μl PBS, sonicated to homogenize the suspension, and then assayed for total triglyceride.
Statistical analysis
Results are expressed as the mean ± SD. Data from two groups were compared using unpaired Student’s t tests. A P value less than 0.05 was considered statistically significant.
Results
Overexpression of S100A16 causes insulin resistance and lipid droplet accumulation in mice
Genotyping of S100A16 transgenic (S100A16Tg/+) mice was performed using standard PCR screening of tail genomic DNA with specific primers (Supplementary Figure S1). The tissue specificity of the transgenic at mRNA levels was determined using Q-PCR (Supplementary Figure S2). The transgene was expressed at high levels in all tissues, and expression was especially high in white adipose tissue (WAT) and liver. S100A16KO/+ was generated for use as a negative control; however, reproductive capacity was limited in these animals and prevented further use as a control. Investigations into the reproductive ability of the animals are continuing in our laboratory. Genotyping of S100A16KO/+ mice was performed using standard PCR screening of tail genomic DNA with specific primers (Supplementary Figure S3).
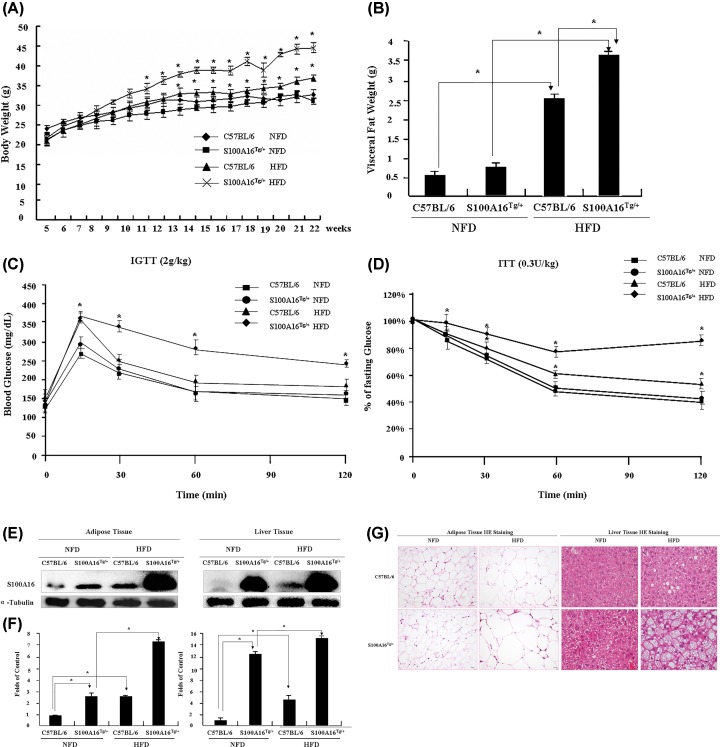
To study the effect of S100A16 overexpression on fat and blood glucose metabolism, the mice (C57BL/6 and S100A16Tg/+ mice) were fed with either a normal fat diet (NFD) or a high fat diet (HFD) for 17 weeks (from 5 to 21 weeks old). The bodyweight was measured every week, and S100A16Tg/+ HFD mice gradually developed a significantly higher body weight than S100A16Tg/+ NFD and C57BL/6 mice. The body weight of C57BL/6 HFD mice was also higher than C57BL/6 NFD mice. There was no difference between S100A16Tg/+ and C57BL/6 NFD groups (Figure 1A). At the experimental end point, the visceral fat was weighed, and visceral fat pad in the HFD groups consistently exceeded that of the NFD groups, with the highest visceral fat weight occurring in S100A16Tg/+ HFD mice (Figure 1B). To assess the impact of S100A16 on glucose metabolism, intraperitoneal glucose tolerance tests (IGTT) and insulin tolerance tests (ITT) were performed during the middle and at the end of feeding. In response to intraperitoneal (IP) glucose infusion (2 g/kg body weight), blood glucose levels were markedly elevated in different groups of mice, with levels in the HFD groups higher than those in NFD mice. However, the elevated blood glucose levels in NFD fed mice (both C57BL/6 and S100A16Tg/+) returned to normal within 120 min. Similarly, blood glucose levels in S100A16Tg/+ HFD mice also decreased, but did not return to normal within 120 min. S100A16Tg/+ mice in the HFD group displayed impaired glucose profiles (Figure 1C). Consistent with the observed glucose intolerance, the response of the blood glucose concentration to an IP injection of insulin was also impaired (Figure 1D). These results demonstrate that overexpression of S100A16 under HFD conditions can result in insulin resistance. After feeding, the expression of S100A16 was evaluated using Western blotting, and the results indicated that S100A16 was overexpressed in adipose tissue (3-fold) and liver tissue (13-fold), and HFD clearly induced S100A16 protein expression in fat tissue (Figure 1E,F). Histological analysis of WAT revealed that the sizes of adipocytes and the lipid droplet accumulation in hepatocytes in S100A16Tg/+ HFD mice were above those in wild-type mice and S100A16Tg/+ NFD mice (Figure 1G). These results demonstrated that overexpression of S100A16 under HFD conditions increased the lipid content in WAT and advanced the fat infiltration in hepatocytes.
Figure 1. Effects of S100A16 on fat metabolism and insulin resistance.
C57BL/6 and S100A16Tg/+ male mice were fed a NFD or HFD from 5 to 21 weeks of age (n = 6 for C57BL/6 NFD mice, n = 5 for S100A16Tg/+ NFD mice, n = 14 for C57BL/6 HFD mice, n = 8 for S100A16Tg/+ HFD mice). (A) Body weight monitored every week. (B) Visceral fat weight measured following administration of the anesthetic Nembutal (100 mg/kg). (C) At 14 and 20 weeks, intraperitoneal glucose tolerance tests (IGTT) were performed twice. All mice were starved overnight, then injected intraperitoneally with glucose at a dose of 2 g/kg of body weight, and blood glucose levels were monitored using a handheld glucometer at 15, 30, 60, and 120 min. (D) The response in blood glucose following an intraperitoneal injection of insulin (0.3 unit/kg of body weight). At 15 and 21 weeks, insulin tolerance tests (ITT) were performed twice. All mice were starved for 12 h, then injected with a bolus of insulin (0.3 unit/kg of body weight), and blood glucose levels were monitored using the handheld glucometer at 15, 30, 60, and 120 min, and blood glucose concentrations at different time points were expressed as a percentage of the fasting blood glucose concentrations. (E) At the end of the study, adipose and liver tissues were removed immediately, and S100A16 protein levels were determined by a Western blotting with an S100A16-specific antibody. (F) Relative expression of S100A16 based on grayscale analysis with α-tubulin as a control. (G) Images of visceral fat cells and liver cells. Adipose and liver tissues were removed, immediately fixed with formalin and embedded in paraffin, and subjected to hematoxylin and eosin (HE) staining (bar = 20 μm).
Effects of S100A16 on gene expression
To further explore the effect of S100A16 by molecular profiling, S100A16Tg/Tg and S100A16KO/+ mouse adipose tissue transcriptomic analyses were performed using the RNA seq as described in the ‘Materials and Methods’ section. Analysis of differentially expressed genes between C57BL/6 and S100A16Tg/Tg mice identified many genes that were up- or down-regulated at least 6-fold in S100A16Tg/Tg adipose tissue (Table 1). Similarly, various genes expression were altered more than 5-fold between C57BL/6 and S100A16KO/+ mice (Table 2).
Table 1. Genes differentially expressed in the adipose tissue of C57BL/6 and S100A16Tg/Tg mice.
| Gene ID | Symbol | log2 ratio (S100A16Tg/Tg/ C57BL/6) | P-value | FDR |
|---|---|---|---|---|
| 84506 | Hamp | 9.523955165 | 5.74E-09 | 1.51E-08 |
| 12501 | Cd3e | 9.397419271 | 4.46E-27 | 2.32E-26 |
| 69496 | Dydc1 | 9.360359767 | 1.55E-18 | 6.19E-18 |
| 114564 | Csprs | 9.359359448 | 2.75E-49 | 2.35E-48 |
| 231932 | Gimap7 | 9.217173553 | 1.31E-21 | 5.77E-21 |
| 170786 | Cd209a | 9.212480404 | 1.10E-24 | 5.33E-24 |
| 328561 | Apol10b | 9.037862288 | 3.97E-29 | 2.18E-28 |
| 106648 | Cyp4f15 | 8.986605799 | 1.71E-30 | 9.73E-30 |
| 433637 | Gm5547 | 8.863585262 | 1.41E-06 | 3.23E-06 |
| 73652 | 2210408F21Rik | 8.763337322 | 1.00E-12 | 3.16E-12 |
| 21935 | Tnfrsf17 | 8.760491769 | 2.47E-10 | 6.98E-10 |
| 622523 | Gm6329 | 8.567368423 | 1.19E-09 | 3.24E-09 |
| 667213 | Gm10416 | 8.53997079 | 2.93E-07 | 6.99E-07 |
| 381484 | Gm5150 | 8.484441432 | 1.19E-09 | 3.24E-09 |
| 93675 | Clec2i | 8.313961722 | 2.61E-09 | 7.00E-09 |
| 320679 | Samd12 | 8.274755981 | 1.26E-08 | 3.26E-08 |
| 241452 | Dhrs9 | 8.177416789 | 2.49E-81 | 3.49E-80 |
| 14990 | H2-M2 | 8.170326953 | 1.00E-12 | 3.16E-12 |
| 93674 | Cml3 | 8.166084423 | 4.06E-15 | 1.42E-14 |
| 72049 | Tnfrsf13c | 8.148405115 | 1.85E-15 | 6.54E-15 |
| 14080 | Fabp1 | 8.135389877 | 0.000159 | 0.00031 |
| 381693 | Wdr95 | 8.122545078 | 1.47E-19 | 6.06E-19 |
| 235712 | Mrgpra2b | 8.074152248 | 5.42E-10 | 1.50E-09 |
| 208154 | Btla | 8.072334743 | 1.49E-75 | 1.94E-74 |
| 631323 | Gm12250 | 8.013441054 | 1.70E-72 | 2.12E-71 |
| 1.01E+08 | 1810008I18Rik | 7.993285786 | 1.50E-05 | 3.18E-05 |
| 83558 | Tex11 | 7.952248761 | 1.23E-22 | 5.62E-22 |
| 16429 | Itln1 | 7.844627104 | 6.08E-08 | 1.51E-07 |
| 98256 | Kmo | 7.815705112 | 3.83E-16 | 1.40E-15 |
| 217306 | Cd300e | 7.808911324 | 1.00E-12 | 3.16E-12 |
| 238722 | Zfp72 | 7.806682871 | 1.12E-10 | 3.23E-10 |
| 68400 | 0610043K17Rik | 7.806441355 | 0.000159 | 0.00031 |
| 209380 | Gm4759 | 7.805974539 | 1.47E-19 | 6.06E-19 |
| 67330 | 1700047M11Rik | 7.781370499 | 7.23E-05 | 0.000146 |
| 414081 | 5330413P13Rik | 7.769601473 | 2.71E-22 | 1.22E-21 |
| 227288 | Cxcr1 | 7.730690992 | 2.93E-07 | 6.99E-07 |
| 236069 | Gm13238 | 7.722722726 | 3.29E-05 | 6.80E-05 |
| 110895 | Slc9a4 | 7.718360701 | 1.23E-22 | 5.62E-22 |
| 11604 | Agrp | 7.717141694 | 3.29E-05 | 6.79E-05 |
| 243958 | Siglecg | 7.697575311 | 8.92E-15 | 3.07E-14 |
| 16175 | Il1a | 7.67156649 | 1.06E-11 | 3.20E-11 |
| 20302 | Ccl3 | 7.667860791 | 3.29E-05 | 6.79E-05 |
| 1.01E+08 | Abhd12b | 7.629120008 | 1.41E-06 | 3.23E-06 |
| 15018 | H2-Q7 | 7.618472764 | 5.74E-09 | 1.51E-08 |
| 18507 | Pax5 | 7.549993026 | 6.44E-07 | 1.50E-06 |
| 320026 | A330076H08Rik | 7.531009923 | 5.42E-10 | 1.50E-09 |
| 74748 | Slamf8 | 7.531009923 | 5.11E-11 | 1.49E-10 |
| 17381 | Mmp12 | 7.525340093 | 1.87E-51 | 1.66E-50 |
| 75526 | Eppin | 7.517571549 | 0.000159 | 0.00031 |
| 383787 | Ankrd63 | 7.500718362 | 1.10E-24 | 5.33E-24 |
| 382045 | Gpr114 | 7.478921412 | 1.96E-14 | 6.64E-14 |
| 76024 | Gm11346 | 7.453565837 | 3.11E-06 | 6.93E-06 |
| 103098 | Slc6a15 | 7.453270173 | 3.41E-18 | 1.34E-17 |
| 353208 | Zfp931 | 7.40658447 | 5.74E-09 | 1.51E-08 |
| 72481 | 2610203C22Rik | 7.40443941 | 1.41E-06 | 3.23E-06 |
| 791403 | D830015G02Rik | 7.384417425 | 0.000159 | 0.00031 |
| 22262 | Uox | 7.355594909 | 5.74E-09 | 1.51E-08 |
| 15930 | Ido1 | 7.337701994 | 1.34E-07 | 3.25E-07 |
| 16631 | Klra13-ps | 7.26974461 | 0.000349 | 0.000662 |
| 70086 | Cysltr2 | 7.262521087 | 2.77E-08 | 7.02E-08 |
| 21949 | Tnfsf8 | 7.251826749 | 5.42E-10 | 1.50E-09 |
| 15551 | Htr1b | 7.244675429 | 1.50E-05 | 3.18E-05 |
| 11889 | Asgr1 | 7.243433333 | 6.82E-06 | 1.48E-05 |
| 637515 | Nlrp1b | 7.240307984 | 7.50E-18 | 2.91E-17 |
| 223917 | Krt79 | 7.235833476 | 5.66E-42 | 4.22E-41 |
| 76074 | Gbp8 | 7.201068057 | 5.86E-41 | 4.27E-40 |
| 1.01E+08 | C230037L18Rik | 7.172216534 | 0.000349 | 0.000662 |
| 78849 | B430010I23Rik | 7.171442888 | 3.63E-79 | 4.96E-78 |
| 68236 | Gtsf1l | 7.159041146 | 0.000349 | 0.000662 |
| 70489 | 5730405O15Rik | 7.147610565 | 0.000349 | 0.000662 |
| 12994 | Csn3 | 7.147249168 | 6.44E-07 | 1.50E-06 |
| 239081 | Tlr11 | 7.139141588 | 4.56E-13 | 1.46E-12 |
| 21947 | Cd40lg | 7.138115307 | 1.50E-05 | 3.18E-05 |
| 232408 | Klrb1f | 7.128308018 | 2.93E-07 | 6.99E-07 |
| 667736 | Gm8787 | 7.084494192 | 6.82E-06 | 1.48E-05 |
| 668108 | Gm8979 | 7.078732515 | 1.41E-37 | 9.54E-37 |
| 140491 | Ppp1r3a | 7.078732515 | 4.27E-74 | 5.44E-73 |
| 232406 | BC035044 | 7.074888505 | 2.47E-10 | 6.98E-10 |
| 94222 | Olig3 | 7.05138094 | 2.77E-08 | 7.02E-08 |
| 1.01E+08 | 4930469G21Rik | 7.042555403 | 0.000349 | 0.000662 |
| 69191 | Pdia2 | 7.039366384 | 6.44E-07 | 1.50E-06 |
| 11997 | Akr1b7 | 7.033510263 | 3.29E-05 | 6.79E-05 |
| 11871 | Art2a-ps | 7.023143548 | 3.29E-05 | 6.79E-05 |
| 66773 | Gm17019 | 7.009909904 | 7.23E-05 | 0.000146 |
| 381287 | A530032D15Rik | 7.007038052 | 6.44E-07 | 1.50E-06 |
| 244550 | Podnl1 | 6.988422374 | 6.08E-08 | 1.51E-07 |
| 18231 | Nxph1 | 6.988306778 | 6.44E-07 | 1.50E-06 |
| 14762 | Gpr33 | 6.971528393 | 1.50E-05 | 3.18E-05 |
| 72958 | Zfp493 | 6.941634158 | 1.06E-11 | 3.20E-11 |
| 236727 | Slc9a7 | 6.933724144 | 5.74E-09 | 1.51E-08 |
| 67717 | Lipf | 6.916966358 | 3.29E-05 | 6.79E-05 |
| 18788 | Serpinb2 | 6.895575376 | 2.93E-07 | 6.99E-07 |
| 58175 | Rgs20 | 6.887153733 | 1.34E-07 | 3.25E-07 |
| 1.01E+08 | AW046200 | 6.877005778 | 1.50E-05 | 3.18E-05 |
| 11835 | Ar | 6.875561879 | 2.47E-10 | 6.98E-10 |
| 210757 | Themis | 6.869010974 | 2.07E-13 | 6.74E-13 |
| 20750 | Spp1 | 6.836426628 | 0 | 0 |
| 382864 | Colq | 6.813142404 | 3.66E-31 | 2.12E-30 |
| 653030 | Arhgap27os3 | 6.788221666 | 5.74E-09 | 1.51E-08 |
| 53315 | Sult1d1 | 6.787333217 | 1.22E-89 | 1.89E-88 |
| 442820 | D830005E20Rik | 6.769849824 | 3.11E-06 | 6.93E-06 |
| 18171 | Nr1i2 | 6.767291601 | 2.77E-08 | 7.02E-08 |
| 54167 | Icos | 6.766348192 | 3.75E-30 | 2.13E-29 |
| 13586 | Ear1 | 6.766348192 | 3.75E-30 | 2.12E-29 |
| 12775 | Ccr7 | 6.734286983 | 1.77E-29 | 9.82E-29 |
| 1E+08 | Cyp4a32 | 6.71609765 | 3.11E-06 | 6.93E-06 |
| 23960 | Oas1g | 6.657746769 | 3.11E-06 | 6.93E-06 |
| 13590 | Lefty1 | 6.655354378 | 3.29E-05 | 6.80E-05 |
| 242248 | Bank1 | 6.633592983 | 1.86E-27 | 9.76E-27 |
| 73895 | 4930431P03Rik | 6.632314689 | 6.44E-07 | 1.50E-06 |
| 269346 | Slc28a2 | 6.616105557 | 4.03E-27 | 2.10E-26 |
| 320046 | F730043M19Rik | 6.604452294 | 7.23E-05 | 0.000146 |
| 226695 | Ifi205 | 6.580481647 | 1.90E-26 | 9.72E-26 |
| 1.01E+08 | 9230112J17Rik | 6.531009923 | 6.44E-07 | 1.50E-06 |
| 620078 | C130026I21Rik | 6.519081102 | 8.10E-74 | 1.03E-72 |
| 433809 | Rnf207 | 6.51285521 | 6.44E-07 | 1.50E-06 |
| 14103 | Fasl | 6.507709835 | 1.50E-05 | 3.18E-05 |
| 19419 | Rasgrp1 | 6.506481066 | 3.77E-49 | 3.21E-48 |
| 16519 | Kcnj3 | 6.49004875 | 7.23E-05 | 0.000146 |
| 78016 | Ccdc150 | 6.487372243 | 9.12E-25 | 4.43E-24 |
| 22445 | Xlr3a | 6.477217106 | 7.23E-05 | 0.000146 |
| 268934 | Grm4 | 6.459081925 | 6.08E-08 | 1.51E-07 |
| 216864 | Mgl2 | 6.448378111 | 3.91E-47 | 3.22E-46 |
| 27278 | Clnk | 6.446727327 | 0.000159 | 0.00031 |
| 12902 | Cr2 | 6.445057088 | 1.85E-15 | 6.55E-15 |
| 15560 | Htr2c | 6.444606385 | 4.83E-12 | 1.48E-11 |
| 11498 | Adam4 | 6.432358525 | 3.11E-06 | 6.93E-06 |
| 108956 | Apol7c | 6.428648205 | 1.50E-05 | 3.19E-05 |
| 209590 | Il23r | 6.425164835 | 1.41E-06 | 3.23E-06 |
| 21822 | Tgtp1 | 6.408300672 | 2.01E-23 | 9.41E-23 |
| 574437 | Xlr3b | 6.40194735 | 0.000159 | 0.00031 |
| 68404 | Nrn1 | 6.401511506 | 9.00E-68 | 1.04E-66 |
| 636104 | Gm7173 | 6.398794728 | 2.93E-07 | 6.99E-07 |
| 20558 | Slfn4 | 6.391267474 | 0 | 0 |
| 382074 | Foxr1 | 6.375731697 | 0.000159 | 0.00031 |
| 1E+08 | Gm14548 | 6.369189971 | 1.50E-05 | 3.18E-05 |
| 244723 | Olfm2 | 6.365807332 | 0.000349 | 0.000661 |
| 14598 | Ggt1 | 6.346016393 | 2.05E-22 | 9.23E-22 |
| 80901 | Cxcr6 | 6.346016393 | 2.05E-22 | 9.23E-22 |
| 15950 | Ifi203 | 6.32032975 | 0 | 0 |
| 234673 | Ces2e | 6.307726344 | 1.41E-06 | 3.23E-06 |
| 217122 | Gm11545 | 6.303456433 | 1.41E-06 | 3.23E-06 |
| 280667 | Adam1b | 6.294542316 | 6.44E-07 | 1.50E-06 |
| 1.01E+08 | Gm13293 | 6.287555886 | 0.000349 | 0.000661 |
| 70928 | Trim69 | 6.279432582 | 0.000159 | 0.00031 |
| 14562 | Gdf3 | 6.276569103 | 3.29E-05 | 6.80E-05 |
| 319940 | Sorbs2os | 6.252733575 | 6.82E-06 | 1.48E-05 |
| 27274 | Zfp354b | 6.24111761 | 6.82E-06 | 1.48E-05 |
| 20981 | Syt3 | 6.223867165 | 6.82E-06 | 1.48E-05 |
| 243659 | Styk1 | 6.218447246 | 1.34E-07 | 3.25E-07 |
| 227485 | Cdh19 | 6.211913419 | 6.44E-07 | 1.50E-06 |
| 226565 | Fmo6 | 6.209081828 | 0.000159 | 0.00031 |
| 399570 | Kank4os | 6.204240565 | 1.50E-05 | 3.18E-05 |
| 13086 | Cyp2a4 | 6.196407998 | 0.000349 | 0.000662 |
| 23845 | Clec5a | 6.18929089 | 4.37E-39 | 3.07E-38 |
| 1E+08 | Dthd1 | 6.174937151 | 1.50E-05 | 3.18E-05 |
| 320692 | 9430037G07Rik | 6.154948391 | 6.44E-07 | 1.50E-06 |
| 17472 | Gbp4 | 6.147617151 | 0 | 0 |
| 667214 | 9930111J21Rik1 | 6.141196601 | 1.40E-92 | 2.26E-91 |
| 239789 | Gmnc | 6.137463105 | 1.26E-08 | 3.26E-08 |
| 170745 | Xpnpep2 | 6.116534547 | 4.59E-19 | 1.86E-18 |
| 320558 | Sycp2 | 6.116534547 | 4.59E-19 | 1.86E-18 |
| 57765 | Tbx21 | 6.108415072 | 1.50E-05 | 3.19E-05 |
| 244233 | Cd163l1 | 6.104043603 | 9.61E-37 | 6.38E-36 |
| 320484 | Rasal3 | 6.091443566 | 2.08E-36 | 1.37E-35 |
| 236312 | Pyhin1 | 6.091443566 | 4.89E-54 | 4.56E-53 |
| 225825 | Cd226 | 6.065908474 | 9.67E-36 | 6.28E-35 |
| 328563 | Apol11b | 6.065908474 | 2.14E-18 | 8.49E-18 |
| 210108 | D130043K22Rik | 6.039913266 | 4.62E-18 | 1.81E-17 |
| 20299 | Ccl22 | 6.039913266 | 4.62E-18 | 1.81E-17 |
| 380842 | Stmnd1 | 6.013441054 | 1.23E-67 | 1.42E-66 |
| 14525 | Gcsam | 6.013441054 | 9.98E-18 | 3.85E-17 |
| 60504 | Il21r | 6.013441054 | 9.98E-18 | 3.85E-17 |
| 13386 | Dlk1 | -15.12358461 | 0 | 0 |
| 14955 | H19 | -10.69854849 | 0 | 0 |
| 21952 | Tnni1 | -10.09528979 | 1.67E-30 | 9.50E-30 |
| 15126 | Hba-x | -8.704567383 | 2.76E-07 | 6.59E-07 |
| 57255 | Cldn13 | -8.250720441 | 7.21E-09 | 1.89E-08 |
| 12824 | Col2a1 | -8.249011178 | 5.28E-40 | 3.77E-39 |
| 27206 | Nrk | -8.202176268 | 1.67E-49 | 1.43E-48 |
| 11609 | Agtr2 | -8.120006692 | 9.64E-21 | 4.13E-20 |
| 23934 | Ly6h | -7.969819615 | 7.21E-09 | 1.89E-08 |
| 228770 | Rspo4 | -7.955994187 | 1.83E-15 | 6.47E-15 |
| 75740 | Egfem1 | -7.82450975 | 5.41E-16 | 1.96E-15 |
| 11606 | Agt | -7.823595489 | 0 | 0 |
| 80982 | Cemip | -7.72334529 | 2.02E-38 | 1.40E-37 |
| 20197 | S100a3 | -7.584625499 | 0.000405 | 0.000763 |
| 14371 | Fzd9 | -7.40435477 | 1.16E-09 | 3.18E-09 |
| 232345 | A2m | -7.355294691 | 5.97E-20 | 2.50E-19 |
| 14840 | Gsg1 | -7.18200142 | 5.76E-06 | 1.26E-05 |
| 16939 | Lor | -7.070501697 | 9.30E-07 | 2.15E-06 |
| 70281 | 2310068J16Rik | -7.036703571 | 0.00022 | 0.000425 |
| 22776 | Zim1 | -6.979851187 | 1.10E-38 | 7.67E-38 |
| 14621 | Gjb4 | -6.950528276 | 3.56E-05 | 7.33E-05 |
| 233332 | Adamts17 | -6.867290381 | 0 | 0 |
| 13717 | Eln | -6.851388429 | 0 | 0 |
| 16682 | Krt4 | -6.815374692 | 9.30E-07 | 2.15E-06 |
| 22044 | Trh | -6.763814261 | 3.30E-33 | 2.01E-32 |
| 619665 | Klf14 | -6.631026918 | 4.46E-08 | 1.11E-07 |
| 71775 | 1300017J02Rik | -6.530196477 | 5.76E-06 | 1.26E-05 |
| 230587 | Glis1 | -6.441055975 | 5.07E-07 | 1.19E-06 |
| 280635 | Emilin3 | -6.310272311 | 8.19E-08 | 2.01E-07 |
| 209966 | Pgbd5 | -6.288340908 | 1.87E-46 | 1.53E-45 |
| 14603 | Gif | -6.225262225 | 2.22E-44 | 1.74E-43 |
| 215303 | Camk1g | -6.222573138 | 6.69E-66 | 7.52E-65 |
| 140494 | Atp6v0a4 | -6.205691868 | 0 | 0 |
| 12291 | Cacna1g | -6.133763871 | 1.57E-41 | 1.15E-40 |
| 626009 | Gm6644 | -6.098998453 | 9.77E-21 | 4.19E-20 |
| 17885 | Myh8 | -6.095711215 | 3.04E-11 | 8.94E-11 |
| 29873 | Cspg5 | -6.081562684 | 2.76E-07 | 6.59E-07 |
| 12839 | Col9a1 | -6.020069209 | 5.07E-07 | 1.19E-06 |
Table 2. Genes differentially expressed in the adipose tissue of C57BL/6 and S100A16KO/+ mice.
| Gene ID | Symbol | log2 ratio (S100A16KO/+/57BL/6) | P-value | FDR |
|---|---|---|---|---|
| 20302 | Ccl3 | 9.775108 | 4.69E-21 | 3.17E-20 |
| 84506 | Hamp | 9.161717 | 1.69E-07 | 5.63E-07 |
| 14080 | Fabp1 | 8.462584 | 9.91E-06 | 2.85E-05 |
| 1E+08 | Gm17455 | 8.356806 | 5.68E-09 | 2.09E-08 |
| 20700 | Serpina1a | 8.151426 | 1.24E-16 | 6.88E-16 |
| 103149 | Upb1 | 7.281212 | 3.33E-07 | 1.08E-06 |
| 218763 | Lrrc3b | 7.250685 | 4.35E-08 | 1.51E-07 |
| 12346 | Car1 | 7.093634 | 1.95E-05 | 5.46E-05 |
| 11699 | Ambp | 7.049678 | 1.95E-05 | 5.46E-05 |
| 22262 | Uox | 6.63972 | 5.03E-06 | 1.48E-05 |
| 67749 | Mgarp | 6.629135 | 7.59E-05 | 0.000199 |
| 75986 | Agmat | 6.447271 | 0.000295 | 0.000722 |
| 268756 | Gulo | 6.259803 | 9.91E-06 | 2.85E-05 |
| 19850 | Rnu3a | 6.127641 | 2.26E-20 | 1.49E-19 |
| 414081 | 5330413P13Rik | 6.076447 | 8.57E-08 | 2.91E-07 |
| 319616 | 5930412G12Rik | 6.045179 | 3.24E-19 | 2.03E-18 |
| 12116 | Bhmt | 5.995014 | 0.00015 | 0.00038 |
| 223780 | Adm2 | 5.972666 | 1.05E-52 | 1.59E-51 |
| 243958 | Siglecg | 5.739626 | 0.00015 | 0.00038 |
| 11287 | Pzp | 5.712604 | 3.47E-15 | 1.81E-14 |
| 78250 | Iqch | 5.596964 | 1.95E-05 | 5.46E-05 |
| 110135 | Fgb | 5.527572 | 1.20E-25 | 9.41E-25 |
| 69121 | Chrdl2 | 5.383981 | 4.89E-12 | 2.18E-11 |
| 14161 | Fga | 5.350034 | 1.66E-22 | 1.17E-21 |
| 72958 | Zfp493 | 5.225759 | 0.000295 | 0.000722 |
| 20704 | Serpina1e | 5.036667 | 6.12E-43 | 7.61E-42 |
| 381813 | Prmt8 | -7.73802 | 8.49E-16 | 4.53E-15 |
| 99738 | Kcnc4 | -7.19525 | 1.21E-13 | 5.81E-13 |
| 22044 | Trh | -7.06465 | 7.17E-39 | 8.11E-38 |
| 52793 | Fam3b | -7.05282 | 0.000409 | 0.000984 |
| 13076 | Cyp1a1 | -6.70754 | 4.93E-09 | 1.82E-08 |
| 58251 | BC100451 | -6.48666 | 5.85E-06 | 1.72E-05 |
| 18741 | Pitx2 | -6.43479 | 1.42E-06 | 4.37E-06 |
| 20608 | Sstr4 | -6.38056 | 0.000409 | 0.000984 |
| 238393 | Serpina3f | -6.30911 | 5.85E-06 | 1.72E-05 |
| 14748 | Gpr3 | -6.28377 | 1.19E-05 | 3.39E-05 |
| 17883 | Myh3 | -6.27053 | 2.93E-14 | 1.45E-13 |
| 69852 | Tcf23 | -6.26505 | 5.89E-10 | 2.32E-09 |
| 96875 | Prg4 | -6.10837 | 2.96E-57 | 4.81E-56 |
| 78789 | Vsig1 | -5.95744 | 1.19E-05 | 3.39E-05 |
| 237310 | Il22ra2 | -5.89815 | 4.89E-05 | 0.000131 |
| 14317 | Ftcd | -5.87517 | 1.57E-32 | 1.50E-31 |
| 268958 | Capn11 | -5.83692 | 4.89E-05 | 0.000131 |
| 12889 | Cplx1 | -5.70496 | 0.000409 | 0.000984 |
| 13166 | Dbh | -5.66327 | 0.000409 | 0.000984 |
| 331374 | Dgkk | -5.65699 | 2.24E-14 | 1.12E-13 |
| 15558 | Htr2a | -5.59687 | 8.87E-14 | 4.30E-13 |
| 56485 | Slc2a5 | -5.46417 | 9.93E-05 | 0.000257 |
| 241520 | Fam171b | -5.4385 | 4.89E-05 | 0.000131 |
| 208188 | Ghsr | -5.41578 | 2.88E-06 | 8.67E-06 |
| 17885 | Myh8 | -5.36421 | 1.08E-11 | 4.72E-11 |
| 76574 | Mfsd2a | -5.29021 | 3.45E-21 | 2.35E-20 |
| 102075 | Plekhg4 | -5.14248 | 4.89E-05 | 0.000131 |
| 20681 | Sox8 | -5.08668 | 1.29E-09 | 4.96E-09 |
| 15116 | Has1 | -5.08668 | 1.29E-09 | 4.95E-09 |
| 13371 | Dio2 | -5.06465 | 6.18E-18 | 3.67E-17 |
| 66198 | Them5 | -5.06465 | 6.18E-18 | 3.67E-17 |
| 246317 | Neto1 | -5.03349 | 0.000409 | 0.000984 |
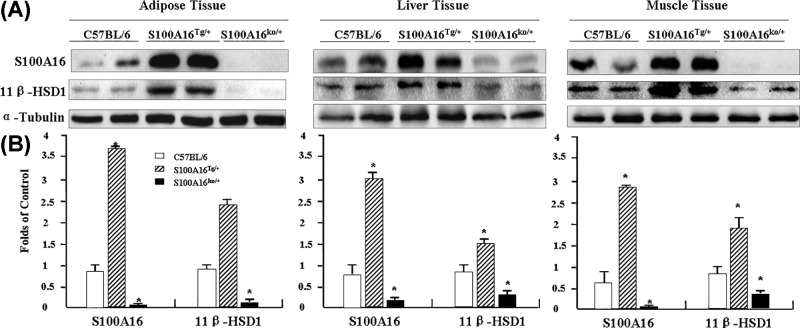
We did not observe any changes in the mRNA expression of 11β-HSD1. However, when we compared 11β-HSD1 protein levels in S100A16Tg/Tg and S100A16KO/+ mice with those in C57BL/6 mice using Western blotting, 11β-HSD1 was up-regulated in S100A16Tg/Tg mice and down-regulated in S100A16KO/+ mice in all these three tissues (visceral adipose, liver, and skeletal muscle from the hind legs) (Figure 2A,B). This difference in S100A16 expression further confirmed that S100A16Tg/Tg and S100A16KO/+ mice had been engineered successfully.
Figure 2. Effect of S100A16 on 11β-HSD1 expression.
Adipose, liver, and muscle tissues were removed from C57BL/6, S100A16Tg/Tg, and S100A16KO/+ mice, and proteins were extracted. (A) Analysis of S100A16 and 11β-HSD1 protein levels by Western blotting. (B) Relative expression of S100A16 and 11β-HSD1 based on grayscale analysis with α-tubulin as a control.
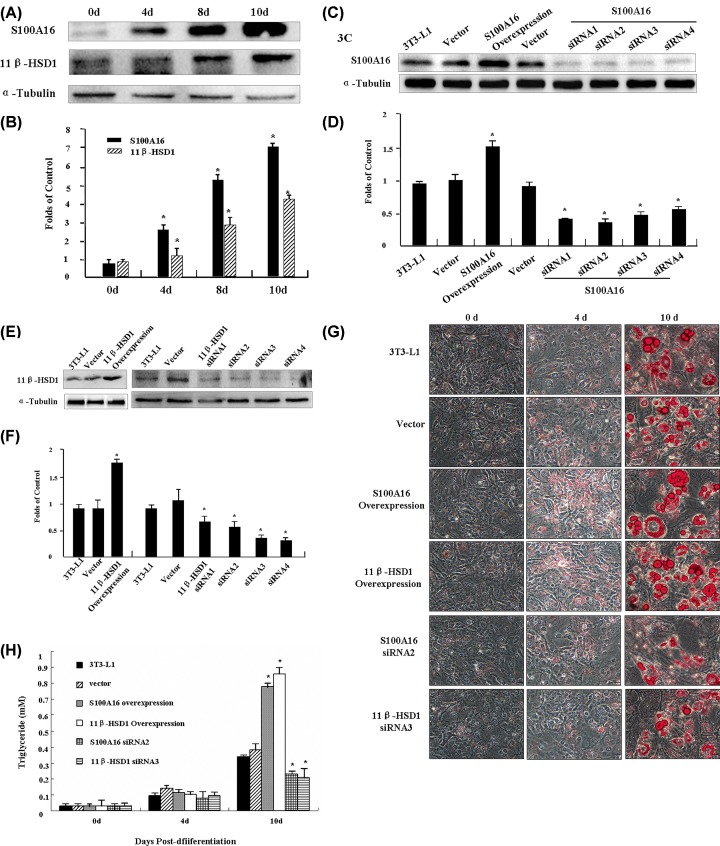
S100A16 and 11β-HSD1 increased lipid droplets in 3T3-L1cells
Expression of S100A16 and 11β-HSD1 at the protein level was measured by Western blotting while 3T3-L1 preadipocytes differentiated into adipocytes. Upon induction of differentiation into adipocytes, expression of both S100A16 and 11β-HSD1 was significantly elevated, by 7.2- and 4.5-fold at 10 days post-differentiation (Figure 3A,B). To assist our understanding of the functional roles of S100A16 and 11β-HSD1 in differentiation, we altered the expression levels in 3T3-L1 preadipocytes using the pcDNA3.1 overexpression system or by transfection with pcDNA6.2-GW/ EmGFP-miR (see ‘Materials and Methods’ section). In either case, we established S100A16-overexpressing and -deficient cell lines and 11β-HSD1-overexpressing and -deficient cell lines for analysis of differentiation (Figure 3C,F).
Figure 3. Effects of S100A16 and 11β-HSD1 on preadipocyte differentiation.
(A) Analysis of S100A16 and 11β-HSD1 protein levels by Western blotting in protein extracts collected at 0, 4, 8, and 10 days after induction of differentiation with α-tubulin as a control. (B) Quantification of S100A16 and 11β-HSD1 expression described in (A) based on grayscale analysis. (C) Western blotting of S100A16 protein levels in various transfectants. (D) Quantification of S100A16 expression described in (C) based on grayscale analysis. (E) Western blotting analysis of 11β-HSD1 protein levels in various transfectants. (F) Quantification of 11β-HSD1 expression described in (E) based on grayscale analysis. (G) Induction of differentiation of 3T3-L1 transfectants overexpressing 11β-HSD1 or S100A16 into adipocytes followed by fixing and staining with oil red O at different time points (0, 4, and 10 days). And induction of differentiation of 3T3-L1 transfectants in which 11β-HSD1 or S100A16 were down-regulated into adipocytes followed by fixing and staining with Oil Red O at different time points (0, 4, and 10 days). Photographs were taken using a light microscope at 200× magnification (LEICA). P ≤0.05 compared to controls. (H) Quantitative determination of triglyceride accumulation in cells. The lipid levels were determined using a Triglyceride GPO-POD assay kit. Results were expressed as the mean±SD for n = 3. P ≤ 0.05 when compared with the corresponding results in the control cell lines.
We investigated whether elevated S100A16 or 11β-HSD1 expression promotes adipogenesis in 3T3-L1 cells harboring different expression vectors by plating them in high density (near confluence) and allowing them to grow for 48 h before initiating differentiation (see ‘Materials and Methods’ section). At different time points after differentiation, Oil Red O staining was applied and microscopy performed to detect cellular lipid droplets. We found that overexpression of S100A16 or 11β-HSD1 led to a marked increase in Oil Red O staining, beginning as early as 10 days after the start of the observation period (Figure 3G), and a reduction in either S100A16 expression (via siRNA2) or 11β-HSD1 expression (via siRNA3) decreased the Oil Red O staining (Figure 3G). Consistent with such staining patterns, quantitative analysis of cellular triglycerides showed that triglyceride accumulation was significantly higher in the cells overexpression of S100A16 and 11β-HSD1 but lower in the cells expression of specific S100A16 siRNA2 and 11β-HSD1 siRNA3, than the control cells (Figure 3H). These results suggest that both S100A16 and 11β-HSD1 promote 3T3-L1 differentiation into adipocytes.
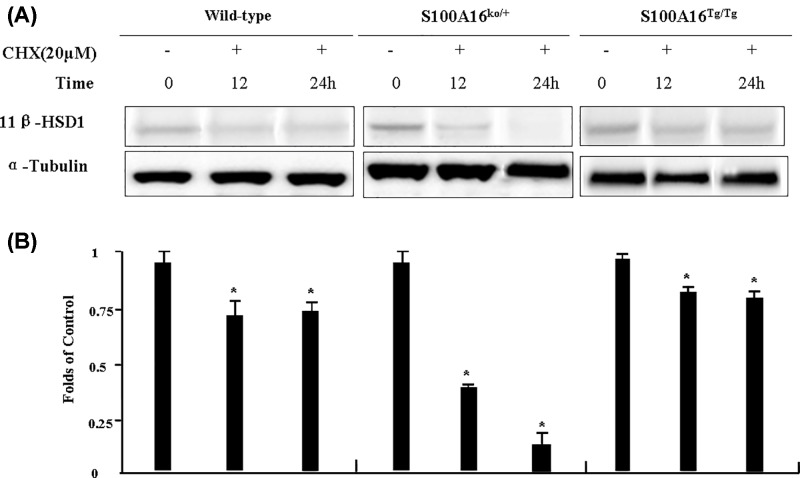
Effects of S100A16 on 11β-HSD1 activity
The above results showed that both S100A16 and 11β-HSD1 have an inductive effect on preadipocyte differentiation, and importantly, S100A16 has a positive influence on 11β-HSD1 protein expression, but the mechanisms remained unknown. We therefore examined numerous molecular signals but the results were not conclusive (data not show). Therefore, we want to investigate whether the stabilization of 11β-HSD1 was affected by S100A16 protein in MEF cells. To study the effect of S100A16 on 11β-HSD1 protein degradation, 20 μM cycloheximide (CHX) was used as a protein synthesis inhibitor. The Western blotting results showed that overexpression of S100A16 inhibited the degradation of 11β-HSD1, and down-regulation of S100A16 had the opposite effect (Figure 4).
Figure 4. Effect of S100A16 on 11β-HSD1 activity.
(A) Mouse embryonic fibroblasts (MEFs) from C57BL/6, S100A16Tg/Tg and S100A16KO/+ mice were treated with cycloheximide (CHX) at 20 μM for different times (0, 12, or 24 h) and cell lysates were subjected to immunoblotting using anti-11β-HSD1 antibody. (B) Quantification of 11β-HSD1 expression described in (A) based on grayscale analysis. *P≤0.05 compared to controls.
Discussion
Our main aims in the present study were (1) to better understand the effect of S100A16 on fat metabolism and insulin resistance, and (2) to gain insight into the mechanism by which S100A16 regulates 11β-HSD1 in adipose tissue. Using a transgenic mouse model fed a NFD or HFD, we investigated the effect of S100A16 on weight gain, visceral fat, insulin sensitivity, and 11β-HSD1 expression. Additionally, using the 3T3-L1 cell model, we investigated the effect of S100A16 and 11β-HSD1 on pre-adipocyte differentiation, and using MEF cells, we investigated the mechanism by which S100A16 up-regulates 11β-HSD1 expression. We showed that overexpression of S100A16 enhanced body weight in HFD mice, and that S100A16 up-regulates 11β-HSD1 protein levels by inhibiting its degradation. These findings are consistent with the ability of 11β-HSD1 to promote preadipocyte differentiation.
S100A16 is a calcium-binding protein that is ubiquitously expressed in human tissues and highly expressed in a variety of tumors [20,21]. Thus, along with many other S100 family members, it may be a potential marker of tumor cells and is likely involved in the regulation of cell growth. However, perturbation of cellular S100A16 resulted in only a modest effect on the proliferation of 3T3-L1 preadipocytes, although it did have a profound impact on differentiation into adipocytes [22,4]. In this context, our present in vivo and in vitro results significantly advance our knowledge by showing that S100A16Tg/Tg mice gradually developed a significantly higher body weight than C57BL/6 mice, and the weight of their visceral fat pads consistently exceeded those of the C57BL/6 group (Figure 1B). Additionally, overexpression of S100A16 led to a marked increase in lipid droplet accumulation as early as 10 days after the start of the observation period while down-regulation of S100A16 expression by siRNA2 had the reverse effect (Figure 3G). RNAseq was done, but no pro-adipogenic markers were identified, which raised the possibility that adipogenesis itself has not been promoted and lipid homeostasis might be disrupted. And this idea need more experiments in futhure.
Obesity is known to increase the risk of metabolic diseases. Increased lipid accumulation in ectopic tissues, including skeletal muscle and liver, is associated with the pathogenesis of non-alcoholic fatty liver disease, cardiac dysfunction, heart failure, and impaired insulin signaling [23–25]. Consistent with this, we showed that overexpression of S100A16 induced lipid accumulation in adipose and liver tissue (Figure 1G), and triggered insulin resistance (Figure 1C,D). Using the intraperitoneal glucose tolerance test, we found that S100A16Tg/+ mice fed a HFD displayed impaired glucose tolerance. Furthermore, consistent with the observed glucose intolerance, the response in blood glucose concentration to an intraperitoneal injection of insulin was also impaired.
11-β-hydroxysteroid dehydrogenase 1 (11β-HSD1) converts cortisone to cortisol, and this enzyme is ubiquitously distributed in glucocorticoid target organs [26]. Up-regulation of 11β-HSD1 is followed by adult obesity and metabolic syndrome, and 11β-HSD1 levels increase with differentiation of mature adipocytes [27]. Therefore, when we found that S100A16Tg/+ mice gradually developed significantly higher body weight than C57BL/6 mice, we decided to measure 11β-HSD1 expression, and found it to be up-regulated in S100A16Tg/Tg mice and down-regulated in S100A16KO/+ mice in fat, liver, and muscle tissue (Figure 2A,B). Additionally, both S100A16 and 11β-HSD1 promoted 3T3-L1 differentiation into adipocytes (Figure 3). These results indicated that S100A16 may be associated with 11β-HSD1. We therefore investigated the effect of S100A16 on gene expression using RNA seq. mRNAs are differentially abundant in C57BL/6 and S100A16Tg/Tg mice. The results revealed that many genes expression were up- or down-regulated at least 6-fold in the adipose tissue of S100A16Tg/Tg mice (Table 1), while other genes expression were altered more than 5-fold between C57BL/6 and S100A16KO/+ mice (Table 2). We did not observe any changes in 11β-HSD1 mRNA levels. These results suggest that the increase in 11β-HSD1 protein levels was due, at least in part, to stabilization against protein degradation. We did not exclude the effect of S100A16 overexpression in other tissues. The phenotype may be a result of gene over expression in the brain to generate an impact on energy balance.
CHX, a well-known inhibitor of protein biosynthesis in eukaryotic cells, binds to the ribosomal E-site and inhibits eukaryotic elongation factor 2 (eEF2)-mediated tRNA translocation [28]. We used 20 μM CHX as a protein synthesis inhibitor to study the effect of S100A16 on 11β-HSD1 protein degradation in MEF cells. The Western blotting results showed that overexpression of S100A16 inhibited the degradation of 11β-HSD1, while down-regulation of S100A16 had the reverse effect (Figure 4). Although the underlying mechanisms remain to be systematically investigated, the initial results described here offer some mechanistic insight into how S100A16 expression impacts 11β-HSD1 activity. There may not be opposite effect between knockdown and trangene of S100A16. Both move in a similar direction however since the tansgene is under the CAG promoter protein expression is no longer regulated by endogenous mechanisms, hence degradation does not occur to a similar extent. Whereas for the knockdown of S100A16, CHX has inhibited protein synthesis to a greater extent. We need more experiments to validate this view.
Conclusions
In conclusion, we showed that overexpression of S100A16 induced preadipocyte differentiation and stimulated body weight gain in mice fed a high fat diet via a mechanism that involves 11β-HSD1.
Supporting information
Supplementary Figure S1. Genotyping of S100A16 transgenic (S100A16Tg/+) mouse using PCR.
Supplementary Figure S2. The levels of S100A16 isoforms in wild-type C57BL/6 and S100A16 transgenic heterzygous (S100A16Tg/+) mice were determined by Q-PCR.
Supplementary Figure S3. Targeted Disruption of the s100a16 Locus.
Supplementary Table S1. Product Data-D12451.
Supplementary Table S2. S100A16 Reverse Complementary Oligonucleotides.
Supplementary Table S3. 11β-HSD1 Reverse Complementary Oligonucleotides.
Abbreviations
- 11β-HSD1
11-β-hydroxysteroid dehydrogenase 1
- HFD
high fat diet
- NFD
normal fat diet
- S16KO/+
S100A16 knockout heterozygous mouse
- S16Tg/+
S100A16 transgenic heterozygous mouse
Funding
The present study was supported by the Natural Science Foundation of Jiangsu Province [BK20141026 DA14]; the National Natural Science Foundation of China [81600001 BJ16 and 81873105]; the Jiangsu Province’s Key Provincial Talents Program [ZDRCA2016005]; the Jiangsu Science & Technology Program [BE2016002-4]; Nanjing Science & Technology Program [201608003]; and the Jiangsu Province Young Medical Talents [QNRC2016598].
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Author Contribution
R.Z., D.L., and X.B. conducted the molecular and the animal studies, carried out the data collection, and wrote this paper; J.K., P.T. and J.Y. conducted the animal studies; L.X. and S.L. conducted part of the molecular studies; and Y.L. designed this study.
References
- 1.Acosta A., Streett S., Kroh M.D., Cheskin L.J., Saunders K.H., Kurian M.. et al. (2017) White Paper AGA: POWER-practice guide on obesity and weight management, education and resources. Clin. Gastroenterol. Hepatol. 30988–30990 S1542-3565 [DOI] [PubMed] [Google Scholar]
- 2.Lauterbach M.A. and Wunderlich F.T. (2017) Macrophage function in obesity-induced inflammation and insulin resistance. Pflugers Arch. 2017, 385–396 10.1007/s00424-017-1955-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Y., Sun Y., Zhu T., Xie Y., Yu J., Sun W.L.. et al. (2007) 11 beta-hydroxysteroid dehydrogenase type 1 promotes differentiation of 3T3-L1 preadipocyte. Acta Pharmacol. Sin. 28, 1198–1204 10.1111/j.1745-7254.2007.00602.x [DOI] [PubMed] [Google Scholar]
- 4.Liu Y., Zhang R.H., Xin J., Sun Y., Li J., Wei D.. et al. (2011) Identification of S100A16 as a novel adipogenesis promoting factor in 3T3-L1 cells. Endocrinology 152, 903–911 10.1210/en.2010-1059 [DOI] [PubMed] [Google Scholar]
- 5.Raajendiran A., Tsiloulis T. and Watt M.J. (2016) Adipose tissue development and the molecular regulation of lipid metabolism. Essays Biochem. 60, 437–450 10.1042/EBC20160042 [DOI] [PubMed] [Google Scholar]
- 6.Marenholz I., Heizmann C.W. and Fritz G. (2004) S100A16, a ubiquitously expressed EF-hand protein which is up-regulated in tumors. Biochem. Biophys. Res. Commun. 313, 237–244 10.1016/j.bbrc.2003.11.115 [DOI] [PubMed] [Google Scholar]
- 7.Li Q., Baines K.J., Gibson P.G. and Wood L.G. (2016) Changes in expression of genes regulating airway inflammation following a high-fat mixed meal in asthmatics. Nutrients 8, E30 10.3390/nu8010030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saito K., Kobayashi M., Nagashio R., Ryuge S., Katono K., Nakashima H.. et al. (2015) S100A16 is a prognostic marker for lung adenocarcinomas. Asian Pac. J. Cancer Prev. 16, 7039–7044 10.7314/APJCP.2015.16.16.7039 [DOI] [PubMed] [Google Scholar]
- 9.Sapkota D., Bruland O., Parajuli H., Osman T.A., Teh M.T., Johannessen A.C.. et al. (2015) S100A16 promotes differentiation and contributes to a less aggressive tumor phenotype in oral squamous cell carcinoma. BMC Cancer 15, 613 10.1186/s12885-015-1622-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M., Ichikawa-T N., Shishito N., Nishiura K., Miura T., Hozumi A.. et al. (2015) Co-expression of S100A14 and S100A16 correlates with a poor prognosis in human breast cancer and promotes cancer cell invasion. BMC Cancer 15, 53. 10.1186/s12885-015-1059-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou W., Pan H., Xia T., Xue J., Cheng L., Fan P.. et al. (2014) Up-regulation of S100A16 expression promotes epithelial -mesenchymal transition via Notch1 pathway in breast cancer. J. Biomed. Sci. 21, 10.1186/s12929-014-0097-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu W., Xue Y., Liang C., Zhang R., Zhang Z., Li H.. et al. (2016) S100A16 promotes cell proliferation and metastasis via AKT and ERK cell signaling pathways in human prostate cancer. Tumour Biol. 37, 12241–12250 10.1007/s13277-016-5096-9 [DOI] [PubMed] [Google Scholar]
- 13.Incollingo Rodriguez A.C., Epel E.S., White M.L., Standen E.C., Seckl J.R. and Tomiyama A.J. (2015) Hypothalamic-pituitary-adrenal axis dysregulation and cortisol activity in obesity: a systematic review. Psychoneuroendocrinology 62, 301–318 10.1016/j.psyneuen.2015.08.014 [DOI] [PubMed] [Google Scholar]
- 14.Chapman K., Holmes M. and Seckl J. (2012) 11b-hydroxysteroid dehydrogenases: intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 93, 1139–1206 10.1152/physrev.00020.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stimson R.H. and Walker B.R. (2013) The role and regulation of 11β-hydroxysteroid dehydrogenase type 1 in obesity and the metabolic syndrome. Horm. Mol. Biol. Clin. Investig. 15, 37–48 [DOI] [PubMed] [Google Scholar]
- 16.Masuzaki H., Tanaka T. and Nakao K. (2004) Novel transgenic mouse model of the metabolic syndrome. Nihon Rinsho. 62, 1059–1065 [PubMed] [Google Scholar]
- 17.Seckl J.R. (2004) 11beta-hydroxysteroid dehydrogenases: changing glucocorticoid action. Curr. Opin. Pharmacol. 4, 597–602 10.1016/j.coph.2004.09.001 [DOI] [PubMed] [Google Scholar]
- 18.Tomlinson J.W., Finney J., Gay C., Hughes B.A., Hughes S.V. and Stewart P.M. (2008) Impaired glucose tolerance and insulin resistance are associated with increased adipose 11beta-hydroxysteroid dehydrogenase type 1 expression and elevated hepatic 5alpha-reductase activity. Diabetes 57, 2652–2660 10.2337/db08-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimba S., Wada T., Hara S. and Tezuka M. (2004) EPAS1 promotes adipose differentiation in 3T3-L1 cells. J. Biol. Chem. 279, 40946–40953 10.1074/jbc.M400840200 [DOI] [PubMed] [Google Scholar]
- 20.Marenholz I., Heizmann C.W. and Fritz G. (2004) S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem. Biophys. Res. Commun. 322, 111122 10.1016/j.bbrc.2004.07.096 [DOI] [PubMed] [Google Scholar]
- 21.Sturchler E., Cox J., Durussel I., Weibel M. and Heizmann C.W. (2006) S100A16, a novel calcium-binding protein of the EF-hand superfamily. J. Biol. Chem. 281, 38905–38917 10.1074/jbc.M605798200 [DOI] [PubMed] [Google Scholar]
- 22.Heizmann C.W. (2002) The multifunctional S100 protein family. Methods Mol. Biol. 172, 69–80 [DOI] [PubMed] [Google Scholar]
- 23.Badin P.M., Langin D. and Moro C. (2016) Dynamics of skeletal muscle lipid pools. Trends Endocrinol. Metab. 24, 607–615 10.1016/j.tem.2013.08.001 [DOI] [PubMed] [Google Scholar]
- 24.Borén J., Taskinen M.R., Olofsson S.O. and Levin M. (2013) Ectopic lipid storage and insulin resistance: a harmful relationship. J. Intern. Med. 274, 25–40 10.1111/joim.12071 [DOI] [PubMed] [Google Scholar]
- 25.Ress C. and Kaser S. (2016) Mechanisms of intrahepatic triglyceride accumulation. World J. Gastroenterol. 22, 1664–1673 10.3748/wjg.v22.i4.1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang Z., Guo C.M., Zhu P., Li W.J., Leslie M. and Sun K. (2007) Role of glucocorticoid receptor and CCAAT/enhancer-binding protein a in the feed-forward induction of 11b-hydroxysteroid dehydrogenase type 1 expression by cortisol in human amnion fibroblasts. J. Endocrinol. 195, 241–253 10.1677/JOE-07-0303 [DOI] [PubMed] [Google Scholar]
- 27.Morton N.M. (2009) Obesity and corticosteroids: 11β-Hydroxysteroid type 1 as a cause and therapeutic target in metabolic disease. Mol. Cell. Endocrinol. 16, 154–164 [DOI] [PubMed] [Google Scholar]
- 28.Schneider-Poetsch T., Ju J., Eyler D.E., Dang Y., Bhat S., Merrick W.C.. et al. (2010) Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat. Chem. Biol. 6, 209–217 10.1038/nchembio.304 [DOI] [PMC free article] [PubMed] [Google Scholar]