Abstract
Serine/arginine (SR)-rich proteins are critical for the regulation of alternative splicing (AS), which generates multiple mRNA isoforms from one gene and provides protein diversity for cell differentiation and tissue development. Genetic evidence suggests that Drosophila genital-specific overexpression of SR-related nuclear matrix protein of 160 kDa (SRm160), an SR protein with a PWI RNA-binding motif, causes defective development only in male flies and results in abnormal male genital structures and abnormal testis. However, the molecular characterization of SRm160 is limited. Using the high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) method in two sex-specific embryonic cell lines, S2 from the male and Kc from the female, we first identified the genome-wide RNA-binding characteristics of SRm160, which preferred binding to the exonic tri-nucleotide repeats GCA and AAC. We then validated this binding through both in vitro gel-shift assay and in vivo splicing of minigenes and found that SRm160 level affects AS of many transcripts. Furthermore, we identified 492 differential binding sites (DBS) of SRm160 varying between the two sex-specific cell lines. Among these DBS-containing genes, splicing factors were highly enriched, including transformer, a key regulator in the sex determination cascade. Analyses of fly mutants demonstrated that the SRm160 level affects AS isoforms of transformer. These findings shed crucial light on SRm160’s RNA-binding specificity and regulation of AS in Drosophila sex determination and development.
Keywords: SR protein, SRm160, HITS-CLIP, RNA-binding motif, Drosophila, sexual development
Introduction
Removal of introns and ligation of exons from nascent RNA transcripts, called pre-mRNA splicing, is an essential step during RNA processing in all eukaryotes (Tollervey and Caceres, 2000; Derrien et al., 2012). The spliceosome is a large and dynamic ribonucleoprotein complex which contains five snRNAs (U1, U2, U4, U5, and U6) and >100 proteins and catalyzes the removal of introns. During spliceosome assembly, snRNA base pairing recognizes intronic sequences including the 5′ splice site (SS), the 3′ SS and the branch site (Will and Luhrmann, 2011; Wan et al., 2016). Alternative splicing (AS) produces multiple mRNA isoforms from one transcript and plays a fundamental regulatory role in cellular differentiation and tissue development (Mohr and Hartmann, 2014; Chen et al., 2015). It occurs when the spliceosome recognizes and uses different SSs, altering the exons included in the RNA product.
AS is regulated through interactions between multiple short cis-acting RNA elements and numerous trans-acting factors including serine/arginine (SR) proteins (Fu and Ares, 2014; Lee and Rio, 2015). The cis-acting RNA elements are classified into splicing enhancers and silencers, which may be located in either exons or introns and are recognized by trans-acting factors. These interactions vary between cell types, during differentiation, and across developmental stages, and result in the AS of exons that produces multiple mRNA isoforms (Shin and Manley, 2002; Black, 2003; Wang and Burge, 2008). Aberrant AS and mutations of SR proteins and other splicing factors are often associated with many human diseases (Cieply and Carstens, 2015; Anczukow and Krainer, 2016).
SR proteins are a super family of proteins with arginine/serine-enriched (RS) domain(s) and RNA recognition motifs (RRM) that have long been recognized to have a major role in the regulation of AS (Long and Caceres, 2009; Shepard and Hertel, 2009 and references therein). SR proteins modulate splicing through various molecular mechanisms including binding of exonic splicing enhancers (ESEs) to improve spliceosomal recognition of weak SSs (Shen et al., 2014), antagonizing splicing inhibitors to increase the usage of SSs (Smith and Valcarcel, 2000), or binding directly to introns to inhibit splicing (Shin and Manley, 2002). Additionally, interactions between multiple SR proteins coordinate the regulation of AS by facilitating initial SS recognition and improving the base pairing between SSs of the nascent RNA transcript and snRNAs (Kohtz et al., 1994; Anko et al., 2012; Pandit et al., 2013; Brooks et al., 2015). Thus, identification of cis RNA-binding motifs on target genes is fundamental to reveal mechanism of SR protein-regulated AS and their role in development.
A variety of systematic approaches have been used to identify putative RNA-binding motifs including in vitro functional systematic evolution of ligands by exponential enrichment (SELEX) (Shin et al., 2004) and recently developed high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) (Licatalosi et al., 2008; Yeo et al., 2009; Darnell, 2010; Jungkamp et al., 2011). The latter and various CLIP protocols have become powerful methods to identify endogenous RNA–protein interactions for a number of SR proteins including SRSF1/ASF/SF2 (Sanford et al., 2008, 2009; ; Pandit et al., 2013), SRSF2 (Pandit et al., 2013), U2AF65/RBM39 (Mai et al., 2016), SRSF3, and SRSF4 (Anko et al., 2012).
SRm160, the SR-related nuclear matrix protein of 160-kDa, is a member of the SR proteins, which consists of a PWI motif for RNA binding at the N-terminus and a long and phosphorylated RS domain at the C-terminus (Eldridge et al., 1999; Szymczyna et al., 2003; Bodenmiller et al., 2008; Zhai et al., 2008). Many biochemical experiments have shown that SRm160 is a splicing co-activator (Blencowe et al., 1998) and associates with other SR proteins, including SRSF4/SRp75, hTra2-β, and SRm300 (Eldridge et al., 1999; Blencowe et al., 2000). SRm160 is concentrated in nuclear speckles and associates with many factors involved in chromatin regulation (Wagner et al., 2003; McCracken et al., 2005), 3′-end processing, and RNA transport (McCracken et al., 2002, 2003; ; Wiegand et al., 2003). SRm160 has been shown to have roles in the differentiation and development of several organisms. In humans, reduction of SRm160 inhibits AS of CD44 exon v5, resulting in a decreased tumor invasiveness (Cheng and Sharp, 2006). In Caenorhabditis elegans, a combined reduction of both SRm160 and SRSF2/SC35 led to production of deficient oocytes (Longman et al., 2001). Lastly in Drosophila melanogaster, ubiquitously reduction of SRm160 results in fly lethality, and tissue-specific overexpression of SRm160 exhibits many developmental defects, including roughed eyes and abnormal male genitals (Fan et al., 2014).
However, despite its role in many development processes, characterization of SRm160 has been limited. The genome-wide in vivo RNA-binding sites and consensus motifs of SRm160 have not been revealed, which has limited our ability to determine which genes are directly regulated by SRm160. To gain a more comprehensive understanding, here we use HITS-CLIP technique and provide global RNA-binding maps of SRm160 in two sex-specific Drosophila cell lines (S2 and Kc), identify and validate their consensus motifs, and reveal that SRm160 regulates AS of its target genes. Lastly, we observe hundreds of differential binding sites (DBS) of SRm160 between the male and female cell lines, including one in transformer, a key sex determination factor. The level of SRm160 affects AS of transformer in flies, thereby demonstrating that SRm160 is involved in Drosophila sex determination and sexual development.
Results
Distribution of reads and peaks from SRm160 HITS-CLIP
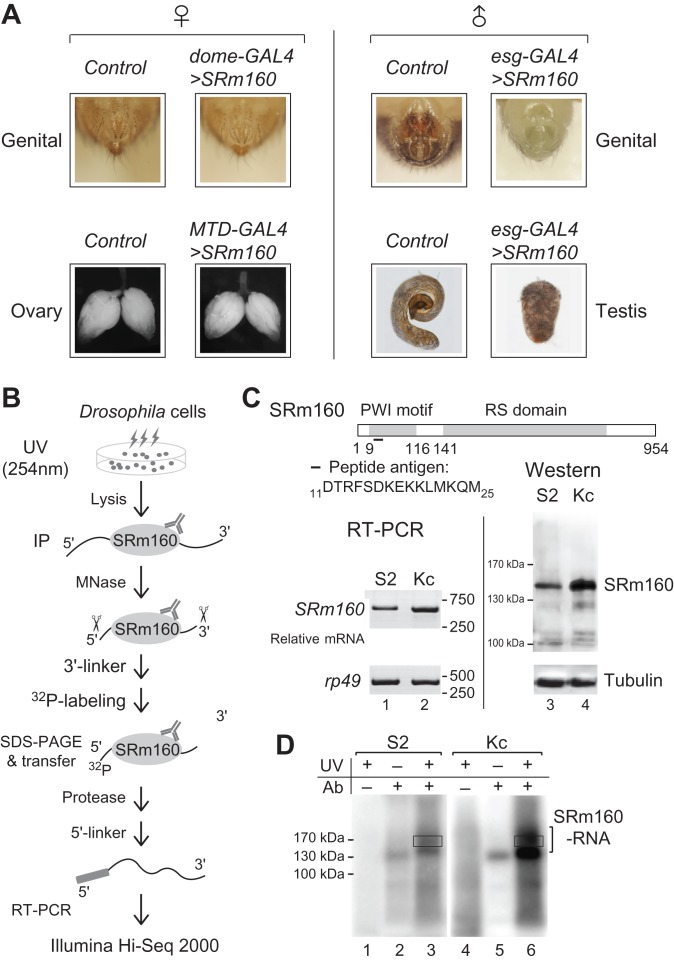
Previous findings have shown that esg-GAL4 driven over-expression (OE) of SRm160 in Drosophila results in defective development of male genital and testis (Fan et al., 2014). To further investigate its effects on females, we used several GAL4 drivers to overexpress SRm160 in female flies, including dome-GAL4 for the female genital (Yan and Perrimon, 2015), MTD-GAL4 for ovary germlines (Ni et al., 2011). The esg-GAL4 driven SRm160-OE flies were used as control and exhibited serious defects in male genital structures and testis development (Figure 1A right), while the male/female ratio was not affected, same as previously described. However, the genital structures and ovary development were not obviously changed in the female flies, in which SRm160-OE was driven by female tissue-specific promoters (Figure 1A left). These results suggested that SRm160-OE affects development of the male reproductive organs but not females, implying that binding to target transcripts of SRm160 would be sexual-different in Drosophila.
Figure 1.
HITS-CLIP of SRm160 in Drosophila sex-specific cell lines. (A) Defective reproduction systems in SRm160-OE Drosophila are only limited in males. Left: female-specific promoters (dome-GAL4 and MTD-GAL4)-induced SRm160-OE did not result in visible defects in female genital and ovary. Right: male-specific promoter esg-GAL4-induced SRm160-OE resulted in lacking of male genital structures and abnormal development of testis. (B) Strategy for SRm160 HITS-CLIP. SRm160-RNA adducts from UV-irradiated Drosophila S2 and Kc cells were purified by D160-1 antibody. RNAs were then partially digested by MNase and ligated with 3′-linker, followed by 32P-labeling. After gel extraction, proteins were digested by Protease K, and remaining RNAs were converted into DNA for illumina sequencing. (C) Schematic of Drosophila SRm160 protein with conserved PWI motif and RS domain. Antigen peptide for antibody D160-1 that was used in immunoprecipitation is indicated. Levels of SRm160 in S2 and Kc cells are detected by RT-PCR (left) and western blot (right). (D) 32P-labeled SRm160-RNA adducts after MNase digestion and 3′-linker ligation. No UV crosslinking or antibody samples are negative controls. Regions for further RNA extraction are indicated in rectangles.
To address this hypothesis, we performed HITS-CLIPs of SRm160 in two Drosophila sex-specific cell lines, S2 and Kc (Figure 1B), which originate from male and female embryos, respectively (Lee et al., 2014). A polyclonal antibody D160-1 was generated against the peptide 11DTRFSDKEKKLMKQM25 of SRm160, which recognized a major band in both cell lines and showed that level of SRm160 in Kc is mildly higher than in S2 (Figure 1C). After co-immunoprecipitation, micrococcal nuclease digestion and 3′-RNA linker ligation, crosslinked SRm160 32P-labeled RNA adducts were observed (Figure 1D), whereas adducts were not present in the samples lacking use of antibody or an unknown no-shift band in the no UV crosslinking controls (Figure 1D and Supplementary Figure S1A). RNAs from the slowly migrating adducts at the range of 170–190 kDa (Figure 1D rectangles) were then isolated for cDNA library construction and Illumina Hi-Seq 2000 sequencing (Supplementary Figure S1B).
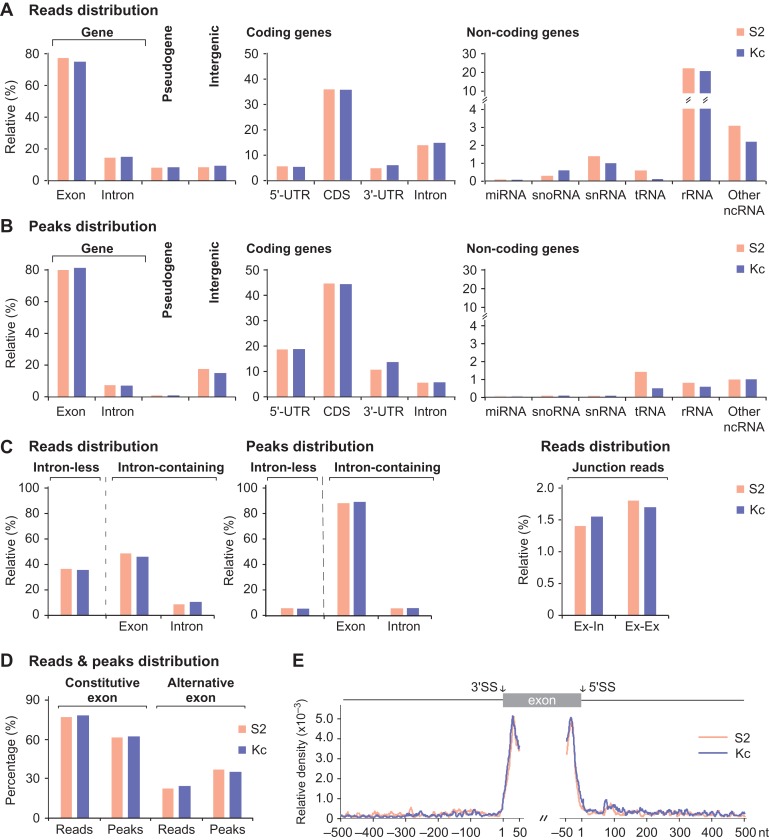
In total, we obtained ~20 M and ~13 M reads from S2 and Kc cells, respectively, in which 8.36 M (S2) and 6.29 M (Kc) reads with an average length of 37-nt were uniquely mapped to the FlyBase reference genome (Supplementary Table S1). Most of the reads were mapped to coding genes, and a small portion of reads mapped to pseudo genes, intergenic regions, and non-coding genes (Figure 2A and Supplementary Table S1). Peaks were called from the clustered reads using MACS (Zhang et al., 2008), and we found 15042 peaks in S2 and 17034 in Kc. Unlike the distribution of reads, peaks were majorly located in exons of coding genes, including regions of 5′-UTR, CDS, and 3′-UTR, and much less located in the introns and non-coding RNAs (Figure 2A and B). Despite the relative large fraction of reads (~20%) are from rRNAs, <0.5% of the total peaks were enriched by those reads (Figure 2A and B), suggesting that they are mostly derived from non-specific background during the CLIP process.
Figure 2.
Genomic landscape of SRm160 HITS-CLIP reads and peaks. (A) Distribution of SRm160 CLIP-seq reads on Drosophila genome, including coding, non-coding, and pseudo genes, and intergenic regions. (B) Distribution of SRm160 peaks on Drosophila genome, which are majorly located in coding-gene regions. (C) CLIP peaks of SRm160 are highly enriched in exons of intron-containing genes. Junction reads analyses indicate that SRm160 binds before and after splicing. Ex–In: exon–intron junction; Ex–Ex: exon–exon junction. (D) Distribution of SRm160 CLIP reads and CLIP peaks on alternative exons and constitutive exons. (E) Statistics of SRm160 CLIP-peak summits. Positions of peak summits are their distance to nearby SSs, and are enriched in both sides of exons. Brown: S2 sample; blue: Kc sample.
In both S2 and Kc samples, ~65% reads were from intron-containing genes and formed >95% of peaks; while in contrast, ~35% reads were from intron-less genes and formed <5% of peaks (Figure 2C and Supplementary Table S1). Considering the roughly equal amounts of intron-less and intron-containing genes in the Drosophila genome (Hoskins et al., 2015), this demonstrates that SRm160 prefers binding of intron-containing genes. To further investigate the effect of SRm160 binding on splicing, we generated a combined library containing the Flybase genome and a library that includes all possible exon–exon junctions within each gene. Reads mapped to exon–exon junctions and exon–intron junctions were at similar levels, around 1.4%–1.8% of total reads (Figure 2C right and Supplementary Table S1 bottom), indicating that SRm160 interacts with both spliced RNA and unspliced precursor RNA. This result is consistent with the previous reports that SRm160 is associated with the EJC complex that functions in mRNA export (McCracken et al., 2003; Wiegand et al., 2003). We also analyzed and compared the distributions of SRm160 CLIP reads and peaks between alternative and constitutive exons and found that SRm160 CLIP reads are distributed in constitutive exons more than in alternative exons. However, the distribution of CLIP peaks in alternative exons is obviously increased, implying that the specificity of SRm160 binding in alternative exons is relatively higher (Figure 2D). In addition, statistical analysis shows that the most frequent positions for SRm160 binding are in exons, 20–40 nts from SSs (Figure 2E). Taken together, SRm160 preferentially binds to exonic sequences of intron-containing genes, consistent with its major function being RNA splicing related. Binding to UTRs and exon–exon junctions confirms that SRm160 remains on the spliced transcripts and may be involved in other RNA processing steps, such as 3′-end processing and RNA transport, as previously described (McCracken et al., 2002, 2003; Wiegand et al., 2003).
Consensus RNA motifs for SRm160 binding
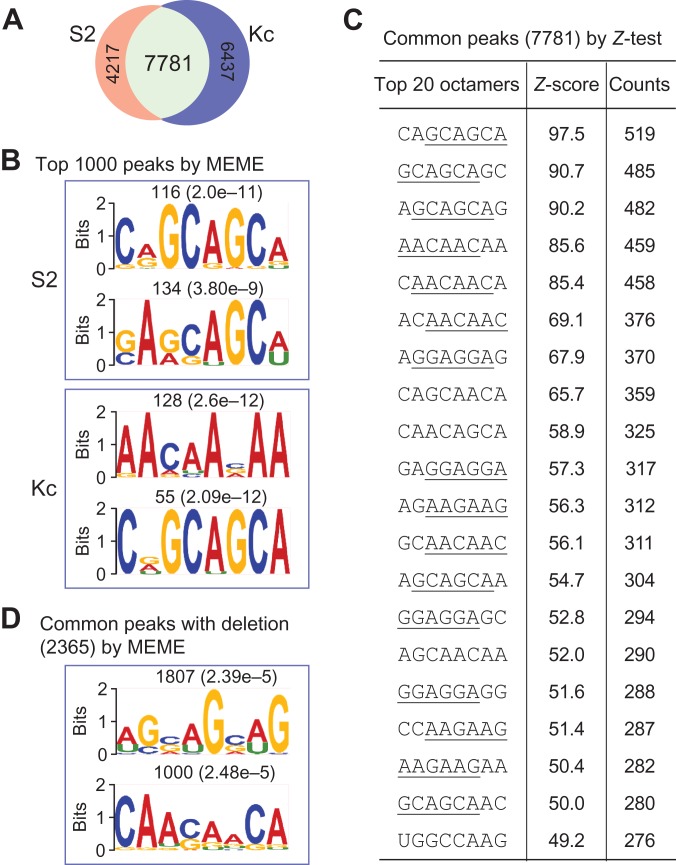
After removing non-reliable peaks from rRNAs, tRNAs, and intergenic regions, there remained 11998 peaks from S2 and 14218 from Kc, in which 7781 peaks are common in both samples (Supplementary Table S2 and Figure 3A). We then searched for consensus RNA motifs in three independent ways.
Figure 3.
Consensus RNA motifs for SRm160 binding. (A) SRm160 CLIP peaks from S2 and Kc samples after removing background bindings. (B) Enriched consensus RNA-binding motifs of SRm160 from the top 1000 peaks in each sample using MEME algorithm. (C) The most enriched 20 octamer sequences of SRm160 binding from common peaks using Z-test analyses. (D) Enriched consensus RNA motifs of SRm160 binding from the deletion-containing peaks using MEME algorithm. These three independent methods generate similar consensus RNA motifs with a common feature of tri-nucleotide repeats, especially GCA and AAC repeats.
First, sequences of the top 1000 peaks from each sample were selected and analyzed by the motif-finding algorithm MEME (Bailey et al., 2009). The most enriched octamers are CAGCAGCA and GAGCAGCA in S2, AACAACAA and CGGCAGCA in Kc, in which three contain GCA repeats and one contains AAC repeats (Figure 3B). Second, we used Z-test to identify over-represented octamer sequences from all and the common peaks (Figure 3C and Supplementary Figure S2). The distribution of octamer sequences from the two samples exhibited high correlations, while no correlation was observed between the peaks and randomly selected genome sequences (Supplementary Figure S2). Similar to the first method, most of the top octamers contain tri-nucleotide repeats, in which GCA and AAC repeats are dominant. Third, we focused on CLIP reads with crosslinking-induced-mutation sites (CIMS) that were generated during the UV crosslinking and reverse transcription (Zhang and Darnell, 2011; Weyn-Vanhentenryck et al., 2014). After removing the SNP-containing reads, we obtained ~2 M reads with mutations of deletions, insertions, and substitutions from each sample (Supplementary Figure S3A) with characteristics consistent with CIMS (Zhang and Darnell, 2011). For example, uridine is the most deleted nucleotide (Supplementary Figure S3B); positions of the deleted nucleotides are predominantly located in the middle region of reads, while insertions and substitutions were not (Supplementary Figure S3C). These characteristics suggest that the deletion-containing reads would provide the most precise location of SRm160 RNA-binding sites. Therefore, we searched sequences from the common peaks containing uridine deletion-reads (2365 peaks) using MEME and found two enriched octamers, AGCAGCAG and CAACAACA, which also contain the GCA and AAC repeats as discovered by our other two methods (Figure 3D).
Taken together, we concluded that the consensus RNA motifs for SRm160 binding in Drosophila are tri-nucleotide repeats and the most frequent repeats are GCA and AAC.
In vitro and in vivo validation of the RNA-binding
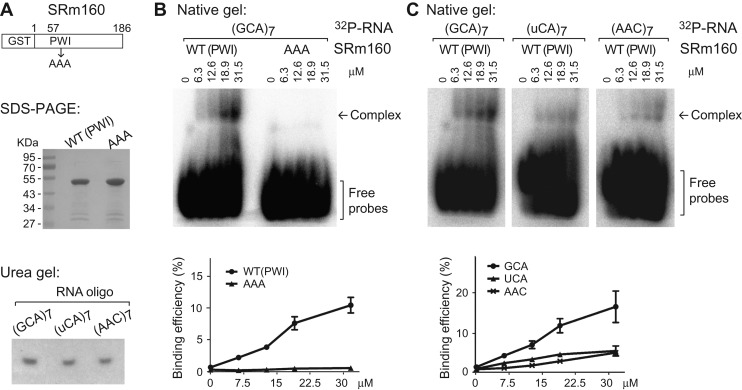
To validate SRm160 binding to the specific tri-nucleotide repeats, we performed in vitro gel-shift assays using purified recombinant WT and mutant SRm160 proteins and 32P-labeled RNA oligonucleotides (Figure 4A). The truncated WT GST-SRm1601−186 (full-length recombinant expression is low) efficiently bound to an RNA oligonucleotides with seven GCA repeats, showing a slowly migrating complex; whereas mutant SRm160 in which the three signature residues (Pro, Trp, and Ile) of the PWI motif were substituted by alanines, totally abolished the RNA-binding ability of SRm160 (Figure 4B). Further, RNA-binding of the WT protein was obviously decreased when the RNA oligonucleotides was switched from GCA repeat to uCA repeat (Figure 4C). These data confirmed that Drosophila SRm160 favors binding to the RNA with GCA repeats, and the binding requires the PWI motif at its N-terminus. We also tested SRm160 binding to the other motif AAC, and found that AAC oligo binding is not as strong as the GCA binding (Figure 4C right). We think this might be to due to additional SRm160 domain is required for AAC binding.
Figure 4.
Gel-shift assays for SRm160 binding with RNA oligonucleotides. (A) Purified GST-SRm1601–186 proteins (coomassie blue staining) and synthesized RNA oligonucleotides (32P-labeled on denaturing Urea-PAGE). (B and C) Native Tris-glycine PAGE for gel-shift assay. Protein concentrations, positions of the RNA–protein complexes, and free RNA probes are indicated. Binding efficiencies are the percentages of RNA–protein complexes to the total probes.
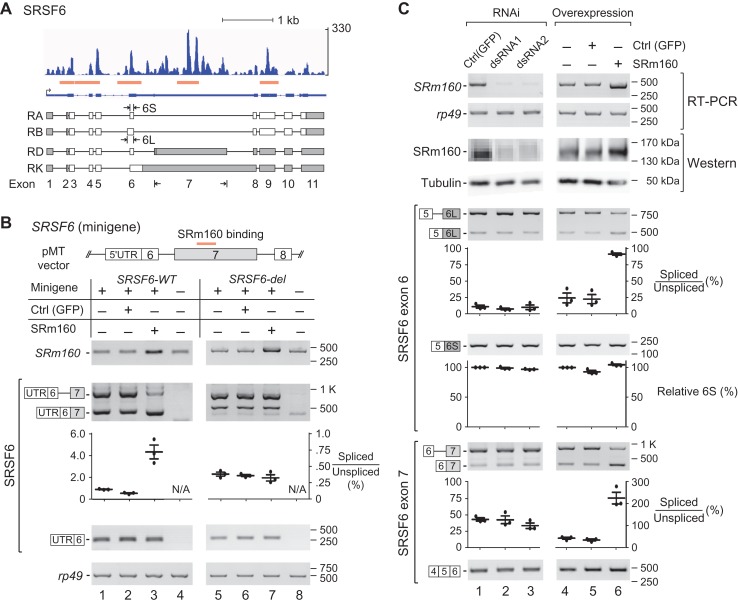
For in vivo validation of the RNA-binding of SRm160, we chose one of the four CLIP peaks on SRSF6/B52 (Figure 5A), which is another SR protein and critical for splicing regulation in Drosophila and mammals (Jensen et al., 2014; Fernando et al., 2015). Minigenes containing SRSF6 exon 7 with the SRm160 CLIP-peak sequence (SRSF6-WT) and without the peak sequence (SRSF6-del) were constructed and transfected into S2 cells. We detected two exon 7-containing isoforms from both minigenes (Figure 5B lanes 1 and 4), and found that splicing of the exon 7 from the WT minigene was enhanced by OE of SRm160 (Figure 5B lane 3). However, for the SRSF6-del minigene that lacks the CLIP-peak sequence, there was no enhanced splicing of exon 7 in the presence of SRm160-OE (Figure 5B lane 7). This in vivo data confirmed that SRm160 binds to the CLIP-peak region in exon 7 of SRSF6.
Figure 5.
SRm160 regulates AS of SRSF6. (A) CLIP peaks of SRm160 on SRSF6 gene. Related AS isoforms of SRSF6 are indicated based on Flybase. Orange boxes: CLIP peaks. (B) AS regulation of SRSF6 is dependent on the SRm160-binding sequence. SRSF6 minigene (WT or deletion) is co-transfected with SRm160-OE (or control) plasmid. (C) AS of exons 6 and 7 in SRSF6 are regulated by SRm160. Upper: SRm160 is either downregulated by dsRNA or upregulated by OE in S2 cells. Two sets of dsRNA and a vector-carried SRm160 CDS are used. Lower: overexpression of SRm160 significantly increases splicing of exons 6 and 7. Semi-quantitation of RT-PCR products from three repeat analyses is presented by scatter plots.
AS regulation by SRm160
Among the four SRm160 CLIP peaks on the SRSF6 transcript, two are located in the alternatively spliced exons 6 and 7 (Figure 5A). Demonstrated by RT-PCR and western blot, we successfully knocked down SRm160 by dsRNA-induced RNAi and over-expressed SRm160 by plasmid transfection in S2 cells (Figure 5C upper). For both exons 6 and 7 in SRSF6, neither of the two sets of different RNAi knockdown of SRm160 changed their AS of SRSF6 (Figure 5C lanes 1–3). In contrast, SRm160-OE significantly changed AS of both exons. SRm160-OE enhanced the usage of an upstream 3′SS and resulted in increased long-isoform of exon 6 (6L); SRm160-OE also increased the usage of the 3′SS of exon 7 resulting in decreased retention of the intron upstream (Figure 5C lanes 4–6), indicating that the SRm160 level has influence on these two AS events. Similarly, SRm160-OE in S2 cells also changed AS patterns of other two tested genes, SRSF1 and SRSF3, their transcripts have SRm160 CLIP peaks (Supplementary Figure S4). Taken together, these results demonstrate that SRm160 level affects AS of its target genes.
DBS of SRm160 in the two sex-specific cells
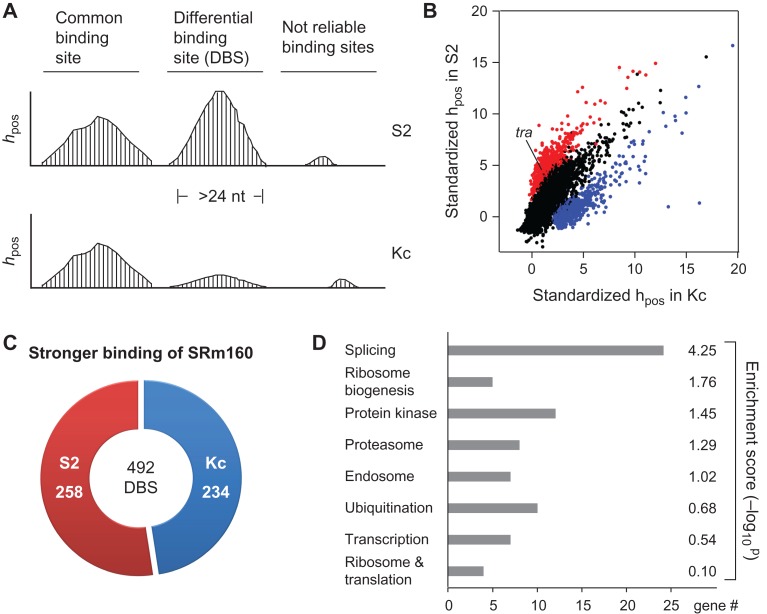
We defined the continuous regions that are longer than 24 nt and with at least one read containing uridine deletion or U-to-C mutation in CLIP peaks as SRm160-binding sites; in total, there are 9188 sites (7072 in Kc and 5385 in Supplementary Tables S2 and S3). To further identify DBS of SRm160 between the two sex-specific cells, we calculated and compared the CLIP reads coverage at each nucleotide position (hpos) in the binding sites (Figure 6A), and found 1138 DBS candidates whose average hpos are significantly different between the two cells (Figure 6B, color dots). After exclusion of the bias expressed genes and unreliable binding sites (details in ‘Materials and methods’ section), we obtained 492 DBSs, in which 234 sites showed stronger SRm160 binding in Kc, while 258 sites showed stronger SRm160 binding in S2 (Supplementary Table S4 and Figure 6C).
Figure 6.
DBS of SRm160 between two sex-specific cell lines. (A) Strategy for identification of SRm160 DBS between Kc and S2 cells. hpos: the CLIP reads coverage at each nucleotide position. (B) Among the total binding sites of SRm160, 1138 are significantly different between Kc and S2. Red: stronger binding sites in S2 cells; blue: stronger binding sites in Kc cells. (C) DBS of SRm160 in Kc and S2 cells. A total of 492 DBSs have been identified. Red: stronger DBSs in S2 cells; blue: stronger DBSs in Kc cells. (D) GO analysis reveals that splicing factors are significantly enriched in the SRm160 DBS-containing genes.
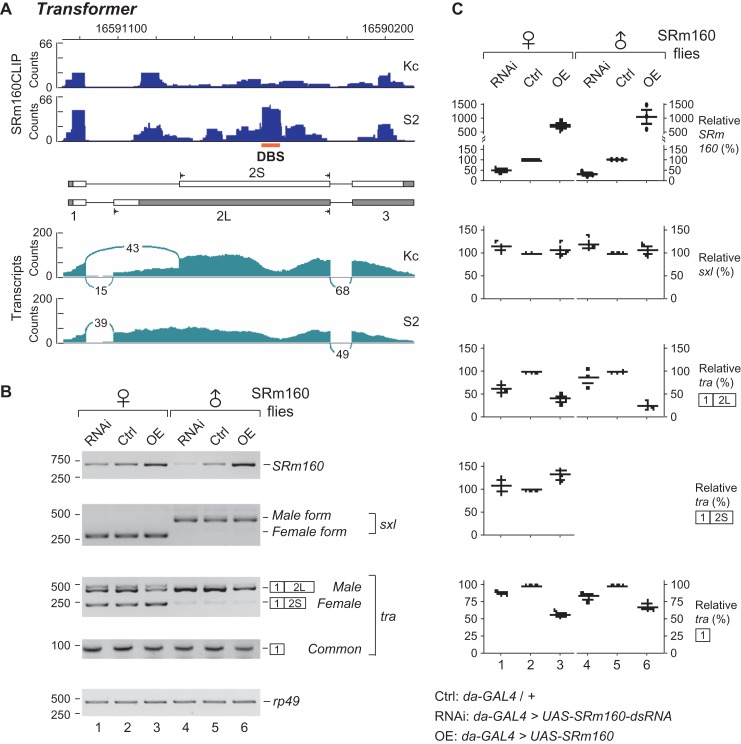
Those 492 DBSs belong to 452 genes, which are significantly enriched in splicing and ribosome biogenesis related pathways through GO analysis (Figure 6D). There are 24 genes of spliceosome components or splicing regulators, including sf3a2, Caper, Prp18, SRSF3, U1 snRNA, U2AF38, and transformer (Supplementary Table S4). In addition, we further investigated sex-specific AS events between S2 and Kc cells. We retrieved raw transcriptome data from the Sequence Read Archive database (SRA) and found 409 AS events that are significantly different between the two sex-specific cell lines (see Methods and Supplementary Table S5), in which 17 AS events having SRm160 DBSs, also including transformer. Therefore, we focused on transformer (tra), a key splicing regulator in the classical Drosophila somatic sex determination pathway (Inoue et al., 1990; Hoshijima et al., 1991). In the exon 2 of tra, there is one SRm160-DBS whose average hpos difference is 37.62 between the Kc and S2 cells, or SRm160 binding to this DBS in S2 cells is ~11 folds strong as in Kc cells (Figure 7A upper). As a control, no SRm160-DBS was found between the Kc and S2 cells on the transcripts of sex lethal (sxl), an upstream gene in the sex determination pathway (Supplementary Figure S5).
Figure 7.
SRm160 regulates AS of transformer. (A) Sex-specific AS of transformer is linked with its differential binding of SRm160. Upper: DBS of SRm160 to the exon 2 of tra is indicated. Lower: transcriptomal analyses indicate that alternative 3′SS of tra exon 2 are sex-specifically selected. (B) AS and expression of transformer are changed in the SRm160 mutant flies. SRm160 level in Drosophila was either downregulated or upregulated by ubiquitous da-GAL4 driver. Isoforms of tra and sxl from the male and female flies were analyzed by RT-PCR. (C) Semi-quantitations of RNA transcripts in the SRm160 flies. RT-PCR products from three repeat analyses are presented by scatter plots. Relative (%): the value of each band is relatively to Ctrl that normalized by rp49 loading control.
In the somatic sex determination pathway, 3′SS of exon 2 in tra is alternatively selected. The upstream 3′SS is used in the male (S2) to generate a single isoform with the long exon 2 (2L); while a downstream 3′SS is a preference shown in the female (Kc) to produce both isoforms, exon 2S (74.1%) and 2L (25.9%) (Figure 7A lower). To address whether SRm160 regulates AS of tra, we investigated SRm160 mutant flies, in which SRm160 is either knocked down by RNAi or overexpressed using the ubiquitously expressed daughterless-GAL4 (da-GAL4) driver. In the SRm160-OE female flies, the isoform 2S was obviously increased and the isoform 2L was significantly decreased in comparison to the control female flies (Figure 7B and C). In the male flies, SRm160-OE decreased the abundance of the single isoform 2L (Figure 7B and C). In addition, using primers to amplify the common exon 1 of tra, we found that overexpression of SRm160 obviously decreased the expression of tra in both males and females (Figure 7B and C bottom). As a control, AS of sxl exhibited no obvious changes in the SRm160 mutant flies. These results demonstrate that upon the SRm160-OE, the transcription is affected in both sexes, and moreover AS of tra have been changed in females, in which the AS regulation would be due to the differential SRm160 binding to the exon 2 of tra and facilitates recognition of the downstream 3′SS. In the SRm160 knockdown flies, we observed less transcription and AS changes of tra than in the SRm160-OE flies (Figure 7).
Discussion
As a member of the SR protein family, SRm160 has a unique PWI RNA binding motif and the longest RS domain of any SR protein (Szymczyna et al., 2003; Bodenmiller et al., 2008). Although SRm160 regulates AS of CD44 in human, contributes to tumor invasiveness (Cheng and Sharp, 2006), and is essential for fly viability and development (Fan et al., 2014), its genome-wide RNA targets, RNA binding motif and how it regulates those genes have remained unclear. Here, we present the analysis of SRm160 binding sites determined by HITS-CLIP, identify its differential binding between two sex-specific Drosophila cells, and have utilized transgenic flies to determine the effect of SRm160 on AS.
The unique PWI motif in SRm160 is located at N-terminus, in which the three signature amino acids (PWI) are 100% conserved across species from fission yeast to humans, while the RS domain containing C-terminus varies significantly in length (Supplementary Figure S6). We prove that the PWI motif is required for RNA binding of Drosophila SRm160, as PWI-to-AAA mutation totally abolishes RNA binding (Figure 4B), consistent with the previous finding of human SRm160 (Szymczyna et al., 2003). The determination of SRm160-binding sites by HITS-CLIP allowed us to use multiple approaches to show that SRm160 binds to many kinds of tri-nucleotide repeats, especially GCA- and AAC- in Drosophila. RNA with GAA repeats has been shown to bind with human SRm160 (Cheng and Sharp, 2006), which is also highly represented in our analysis of enriched octamers for SRm160 binding (Figure 3C). Therefore, to some extent, the RNA motifs for SRm160 binding are conserved in Drosophila and human.
We totally identified 9188 RNA-binding sites of SRm160 in the two sex-specific Drosophila cell lines. The RNA-binding site of SRm160 is defined from a CLIP-peak that contains at least one CIMS-read; this definition makes the binding more reliable.
SRm160 was identified as a splicing co-activator and has been suggested to be involved in several steps of pre-mRNA processing (Eldridge et al., 1999; McCracken et al., 2002; Wagner et al., 2004). Our HITS-CLIP data reveal that SRm160 predominantly binds to exonic sequences in intron-containing genes, suggesting that the major function of SRm160 is RNA splicing related. For most of the AS events tested in this study, SRm160 functions as an activator to enhance recognition of nearby SSs and promote splicing. For example, SRm160-OE facilitates the recognition of the relatively weak 3′SS of exon 7 in SRSF6 (AUGUGCUUUGACUG/A) and the nearby 3′SS of exon 2 in transformer, and thus enhances their splicing (Figures 5B and 7B).
Recently, interactions between RNA-binding proteins and among SR proteins have been widely studied, showing interaction networks that control gene expression and regulation (Anko et al., 2012; Brooks et al., 2015). We identify alternative exons from three SR proteins (SRSF1, SRSF3, and SRSF6) that are bound with and regulated by SRm160; the level of SRm160 significantly affects selection of those alternative exons. The findings that many SR proteins and splicing factors are targets of SRm160 and their AS are regulated by SRm160 lead to a conclusion that SRm160 is an important regulator of cross-talks between SR proteins and splicing factors, regulating the expression and function of other RNA-binding proteins. This provides an SRm160-involved interaction network, which would be critical for the fly development. However, since the interaction network between SR proteins (here SRm160 level regulates AS of three SR proteins), we cannot rule out the possibility that the changed AS events are indirect through alteration of other factors.
In addition, we also tested AS events of four non-SR protein genes (CG10433, CG18659, dre4, and nija) in the SRm160 mutant flies. Transcripts of these four genes have SRm160 DBSs. Our data indicated that AS of three genes (CG18659, dre4, and nija) have been changed in SRm160 mutant flies (Supplementary Figure S7).
We previously found that SRm160-OE in Drosophila causes roughened eyes with severe defects of retinal cells and disordered structures of male genital (Fan et al., 2014). Here, we show strong evidence that SRm160 is involved in the Drosophila sex determination pathway and sexual development. First, we prove that SRm160-OE significantly disrupts the reproductive systems in male flies but not females, indicating that the sexual development of male flies is more sensitive to the level of SRm160. Second, SRm160 has as many as 492 sex-differential RNA-binding sites, many of which are in splicing-related genes, including one site in the important sex determination factor tra. Third, through analysis of SRm160 mutant flies, we find AS of exon 2 in tra is regulated by SRm160. Sex determination pathway in Drosophila has provided many classical examples of AS regulation. A cascade of genes, including sxl, tra, and dsx, are alternatively spliced in sex-specific manners. The upstream SXL protein binds to the intron 1 of tra and inhibits usage of the upstream 3′SS in female flies (Black, 2003). Our finding provides an additional AS regulation of tra in Drosophila. In the future, it would be interesting to explore how SRm160 binds more efficiently to the DBS region on tra in S2 cells with relatively lower expression level than in Kc cells; and to investigate which gene(s) regulated by SRm160 eventually cause(s) the sexual-developmental defects in male flies.
Materials and methods
Cell culture, strains, and plasmids
S2 and Kc cells from D. melanogaster were cultured in the complete Schneider’s Medium (GIBCO) that contains 10% or 5% heat-inactivated FBS (GIBCO). SRm160 CDS was amplified from Drosophila cDNA, the WT SRSF6 minigene containing exons 6–8 was amplified from the fly genomic DNA, and the SRSF6-deletion minigene was constructed, in which a 267-nt fragment in exon 7 was excluded; PCR products were cloned into a pMT-MCS-Flag vector (Yang et al., 2013). Construction primers are listed in the Supplementary Table S6. Culture and crosses of D. melanogaster were carried out on standard medium and at 25°C except as noted. Fly strains include da-GAL4, GMR-GAL4, MTD-GAL4, esg-GAL4, UAS-SRm160 (BDSC), UAS-SRm160-dsRNA (VDRC, 100751), and domeless-GAL4 (Owusu-Ansah and Banerjee, 2009).
Antibodies
Antiserum (D160-1) against Drosophila SRm160 was generated by immunizing rabbits with the peptide 11DTRFSDKEKKLMKQM25. Western blot of anti-tubulin was probed using mAb DM1A (Sigma).
RNAi knockdown and OE of SRm160
In cells, SRm160 knockdown by RNAi was performed with 15 μg/well of non-overlapping dsRNA that specifically targeting to SRm160 for 60 h. All dsRNAs were generated by in vitro transcription using T7 RNA polymerase (Promega). OEs were performed in S2 cells with 1.0 μg/well of each construct using Effectene® Transfection Reagent (QIAGEN) and induced by 0.5 mM CuSO4 for 60 h (Yang et al., 2013). Specificity and efficiency were monitored by RT-PCR and western blot; related primers are listed in the Supplementary Table S6. In fly, to knockdown and to overexpress SRm160, ubiquitous driver da-GAL4 were crossed, respectively, with RNAi strain (VDRC# 100751) and SRm160G18603 (BDSC#26938) at 23°C. Fresh adult flies (0–4 h after eclosion) were collected for further experiments.
RT-PCR
Total RNAs were isolated from the Drosophila cells and adults by TRIzol (Ambion), and examined in quality and concentration after DNase I treatment. RT-PCR was then carried out by RevertAid Reverse Transcriptase (Thermo) and Ex-Taq (TaKaRa). The density of each amplified band was selected and quantitated by Gel-pro Analyzer. Obtained data from three individual repeats were then analyzed and normalized by controls.
HITS-CLIP
A 10-cm dish of S2 cells were harvested and resuspended in 5 ml PBS buffer and irradiated at 254 nm with 400 mJ/cm2 by a CL-1000 Ultraviolet Crosslinker (UVP). The lysate was prepared (Licatalosi et al., 2008) and incubated with antibody D160-1 that coupled to Protein A beads (Millipore). Following procedures are modified based on Yeo et al. (2009). Briefly, after MNase digestion (0.4 units/ml), 3′-RNA linker ligation and 32P-labeling, the SRm160-RNA adducts were separated by a 4%–12% NuPAGE Bis-Tris gel (Invitrogen), transferred to nitrocellulose membrane (Millipore), excised, and treated with protease K (TaKaRa). Recovered RNAs were then ligated with a 5′-RNA linker, reverse transcribed, amplified with DNA adaptors, and paired-end sequenced by Illumina Hi-Seq 2000. RNA linkers and DNA adaptors are listed in the Supplementary Table S6.
Reads and peaks analyses of HITS-CLIP
Raw reads were firstly processed by trimming of adaptors and low-quality bases by Seqtk v1.0 (https://github.com/lh3/seqtk), paired reads were then joined by ea-utils v1.1.1 (https://expressionanalysis.github.io/ea-utils/). Successfully joined reads were defined as effective reads and mapped to the reference genome of D. melanogaster (FlyBase, Release 6.02) using BWA v0.7.10 with default parameters (Li and Durbin, 2010). All mapped reads from this study have been submitted to the SRA database in NCBI under the accession numbers SRS1851748 and SRS1851749. Peaks were subsequently called by MACS v1.4.2 from uniquely mapping reads (Zhang et al., 2008). Distribution was identified in four hierarchical categories based on the annotations from FlyBase. To investigate the splicing-related reads, the processed reads were mapped to the fly genome and an extra exon–exon junction library, which contains all possible junctions between any pair of exons within each gene using a custom Perl script. To calculate the CLIP-peak density relative to SSs, we limited all peaks to 9-nt windows (summits are the central points) and counted each nucleotide by position in the exon or intron. Intronic peaks with >500 nt distance and exonic peaks with >50 nt to any SS were not counted.
Consensus RNA motif searching
Three independent methods were applied. First, MEME7, version 4.4.0 (Bailey et al., 2009) was used to identify enriched discriminative motifs from the P-value ranked top 1000 CLIP-peak sequences in each sample (-mod zoops -minw 6 -maxw 8). Second, the Z-test statistic analysis was performed to calculate the distribution pattern of hexamers, which were then sorted on Z-scores and clustered according to their similarities. Third, according to a recently described pipeline (Zhang and Darnell, 2011), we identified the significantly and frequently mutated sites (insertions, deletions, and substitutions) based on permutation test after ruling out SNPs, and then applied sequences from the deletion CIMS-containing peaks for searching RNA motifs using MEME algorithm.
Gel-shift assay
SRm160 proteins were expressed in Escherichia coli and purified using Glutathione-Sepharose (GE) followed by dialysis against buffer D (Shao et al., 2012). RNA probes were synthesized by TaKaRa and 32P-labeled using T4 DNA ligase (TaKaRa). For gel-shift assay, 0.15–0.5 nmol of proteins were incubated with 10 fmole RNA probe for 30 min at 25°C in a 20-μl mixture with 10 mM HEPES-KOH (pH7.3), 20 mM KCl, 1 mM MgCl2, 1 mM DTT, 100 ng/μl yeast tRNA, and 0.5 mM ATP. Above mixtures were then loaded to a 5% acrylamide gel and run in 0.5× Tris-glycine buffer (25 mM Tris, 50 mM glycine) at 4°C (Das and Reed, 1999) and the gel was exposed by Phosphor Imager (Multi Gauge V3.0).
Analysis of DBS
To find DBS of SRm160 between Kc and S2 cells, we first calculated CLIP reads accumulation that are normalized by total mapped reads on each nucleotide position (hpos), and then compared their differences (dpos) between S2 and Kc samples. Then positions with significant dpos (P < 0.05) were selected. Region with ≥25 continuous significant positions was considered as a DBS candidate. Four extra filters were used to obtain reliable DBSs: (i) should be located in a CLIP-peak and covered by at least one uridine deletion or U-to-C substitution read; (ii) expression level change of belonging gene should be <2 folds between Kc and S2 cells, expressions are calculated from SRA transcriptomal data (SRR1197456 and SRR3040054 for Kc, SRR1197282 and SRR3042565 for S2); (iii) ratio of average hpos change between Kc and S2 should be <0.5 or >3.0 (SRm160 level is slightly higher in Kc); (iv) average dpos should be >25 or ≤25 (if one average hpos is <3, the average dpos is then allowed to be 20). Gene ontology and enrichment were analyzed by DAVID (Huang da et al., 2009).
AS analyses
Raw transcriptome data of Kc cells (SRR3040054 and SRR3040509) and S2 cells (SRR1197282 and SRR3042565) were retrieved from the SRA database, and mapped to the Flybase using HISAT2 under default parameters (Kim et al., 2015). Gene expression levels were calculated by cufflinks (Trapnell et al., 2012) and the splicing changes between Kc and S2 were analyzed by rMATS (Shen et al., 2014). Percentage of splicing in (PSI, Ψ) was calculated with JunctionCountOnly model. Thresholds for significant AS changes are: ΔΨ > 0.1, P-value < 0.05, and FDR < 0.1.
Supplementary Material
Acknowledgements
We thank Charles C. Query and Brian Kosmyna at Albert Einstein College of Medicine for critical reading and discussion, and members in Xu lab for data entries and discussions.
Funding
This work was supported by grants from National Natural Science Foundation of China (NSFC; 31525022, 91440109, and 31472045) and Chinese Academy of Sciences (KJZD-EW-L12) to Y.-Z.X., NSFC (31570821) to Y.-J.F., and NSFC (31522053, 91631103) to S. Z.
Conflict of interest
none declared.
Author contributions
C.Q. and Y.-J.F. contributed to the project initiation, C.Q. contributed to HITS-CLIP and validation, Y.Z. and T.-L.P. contributed to bioinformatics under supervision by S.Z. Y.-J.F. and Y.S. contributed to fly work, Y.-Z.X. contributed to project supervision and manuscript writing. All authors helped to interpret data and edit the manuscript.
References
- Anczukow O., and Krainer A.R. (2016). Splicing-factor alterations in cancers. RNA 22, 1285–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anko M.L., Muller-McNicoll M., Brandl H., et al. (2012). The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 13, R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L., Boden M., Buske F.A., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.L. (2003). Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72, 291–336. [DOI] [PubMed] [Google Scholar]
- Blencowe B.J., Bauren G., Eldridge A.G., et al. (2000). The SRm160/300 splicing coactivator subunits. RNA 6, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J., Issner R., Nickerson J.A., et al. (1998). A coactivator of pre-mRNA splicing. Genes Dev. 12, 996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenmiller B., Campbell D., Gerrits B., et al. (2008). PhosphoPep—a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 26, 1339–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks A.N., Duff M.O., May G., et al. (2015). Regulation of alternative splicing in Drosophila by 56 RNA binding proteins. Genome Res. 25, 1771–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Dai X., and Wu J. (2015). Alternative splicing: an important mechanism in stem cell biology. World J. Stem Cells 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C., and Sharp P.A. (2006). Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell. Biol. 26, 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieply B., and Carstens R.P. (2015). Functional roles of alternative splicing factors in human disease. Wiley Interdiscip. Rev. RNA 6, 311–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell R.B. (2010). HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley interdisciplinary reviews. RNA 1, 266–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R., and Reed R. (1999). Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA 5, 1504–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T., Johnson R., Bussotti G., et al. (2012). The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge A.G., Li Y., Sharp P.A., et al. (1999). The SRm160/300 splicing coactivator is required for exon-enhancer function. Proc. Natl Acad. Sci. USA 96, 6125–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y.J., Gittis A.H., Juge F., et al. (2014). Multifunctional RNA processing protein SRm160 induces apoptosis and regulates eye and genital development in Drosophila. Genetics 197, 1251–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernando C., Audibert A., Simon F., et al. (2015). A role for the serine/arginine-rich (SR) protein B52/SRSF6 in cell growth and myc expression in Drosophila. Genetics 199, 1201–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X.D., and Ares M. Jr. (2014). Context-dependent control of alternative splicing by RNA-binding proteins. Nat. Rev. Genet. 15, 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., et al. (1991). Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252, 833–836. [DOI] [PubMed] [Google Scholar]
- Hoskins R.A., Carlson J.W., Wan K.H., et al. (2015). The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B.T., and Lempicki R.A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hoshijima K., Sakamoto H., et al. (1990). Binding of the Drosophila sex-lethal gene product to the alternative splice site of transformer primary transcript. Nature 344, 461–463. [DOI] [PubMed] [Google Scholar]
- Jensen M.A., Wilkinson J.E., and Krainer A.R. (2014). Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat. Struct. Mol. Biol. 21, 189–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungkamp A.C., Stoeckius M., Mecenas D., et al. (2011). In vivo and transcriptome-wide identification of RNA binding protein target sites. Mol. Cell 44, 828–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Langmead B., and Salzberg S.L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohtz J.D., Jamison S.F., Will C.L., et al. (1994). Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature 368, 119–124. [DOI] [PubMed] [Google Scholar]
- Lee H., McManus C.J., Cho D.Y., et al. (2014). DNA copy number evolution in Drosophila cell lines. Genome Biol. 15, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., and Rio D.C. (2015). Mechanisms and regulation of alternative Pre-mRNA splicing. Annu. Rev. Biochem. 84, 291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi D.D., Mele A., Fak J.J., et al. (2008). HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456, 464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.C., and Caceres J.F. (2009). The SR protein family of splicing factors: master regulators of gene expression. Biochem. J. 417, 15–27. [DOI] [PubMed] [Google Scholar]
- Longman D., McGarvey T., McCracken S., et al. (2001). Multiple interactions between SRm160 and SR family proteins in enhancer-dependent splicing and development of C. elegans. Curr. Biol. 11, 1923–1933. [DOI] [PubMed] [Google Scholar]
- Mai S., Qu X., Li P., et al. (2016). Global regulation of alternative RNA splicing by the SR-rich protein RBM39. Biochim. Biophys. Acta 1859, 1014–1024. [DOI] [PubMed] [Google Scholar]
- McCracken S., Lambermon M., and Blencowe B.J. (2002). SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol. Cell. Biol. 22, 148–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S., Longman D., Johnstone I.L., et al. (2003). An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J. Biol. Chem. 278, 44153–44160. [DOI] [PubMed] [Google Scholar]
- McCracken S., Longman D., Marcon E., et al. (2005). Proteomic analysis of SRm160-containing complexes reveals a conserved association with cohesin. J. Biol. Chem. 280, 42227–42236. [DOI] [PubMed] [Google Scholar]
- Mohr C., and Hartmann B. (2014). Alternative splicing in Drosophila neuronal development. J. Neurogenet. 28, 199–215. [DOI] [PubMed] [Google Scholar]
- Ni J.Q., Zhou R., Czech B., et al. (2011). A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods 8, 405–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., and Banerjee U. (2009). Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature 461, 537–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit S., Zhou Y., Shiue L., et al. (2013). Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol. Cell 50, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Coutinho P., Hackett J.A., et al. (2008). Identification of nuclear and cytoplasmic mRNA targets for the shuttling protein SF2/ASF. PLoS One 3, e3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford J.R., Wang X., Mort M., et al. (2009). Splicing factor SFRS1 recognizes a functionally diverse landscape of RNA transcripts. Genome Res. 19, 381–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W., Kim H.S., Cao Y., et al. (2012). A U1-U2 snRNP interaction network during intron definition. Mol. Cell. Biol. 32, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S., Park J.W., Lu Z.X., et al. (2014). rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. USA 111, E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard P.J., and Hertel K.J. (2009). The SR protein family. Genome Biol. 10, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C., Feng Y., and Manley J.L. (2004). Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature 427, 553–558. [DOI] [PubMed] [Google Scholar]
- Shin C., and Manley J.L. (2002). The SR protein SRp38 represses splicing in M phase cells. Cell 111, 407–417. [DOI] [PubMed] [Google Scholar]
- Smith C.W., and Valcarcel J. (2000). Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25, 381–388. [DOI] [PubMed] [Google Scholar]
- Szymczyna B.R., Bowman J., McCracken S., et al. (2003). Structure and function of the PWI motif: a novel nucleic acid-binding domain that facilitates pre-mRNA processing. Genes Dev. 17, 461–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., and Caceres J.F. (2000). RNA processing marches on. Cell 103, 703–709. [DOI] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Chiosea S., Ivshina M., et al. (2004). In vitro FRAP reveals the ATP-dependent nuclear mobilization of the exon junction complex protein SRm160. J. Cell Biol. 164, 843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S., Chiosea S., and Nickerson J.A. (2003). The spatial targeting and nuclear matrix binding domains of SRm160. Proc. Natl Acad. Sci. USA 100, 3269–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan R., Yan C., Bai R., et al. (2016). Structure of a yeast catalytic step I spliceosome at 3.4 A resolution. Science 353, 895–904. [DOI] [PubMed] [Google Scholar]
- Wang Z., and Burge C.B. (2008). Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14, 802–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck S.M., Mele A., Yan Q., et al. (2014). HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 6, 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H.L., Lu S., and Cullen B.R. (2003). Exon junction complexes mediate the enhancing effect of splicing on mRNA expression. Proc. Natl Acad. Sci. USA 100, 11327–11332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will C.L., and Luhrmann R. (2011). Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, pii: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., and Perrimon N. (2015). spenito is required for sex determination in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 112, 11606–11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wang X.Y., Zhang Z.M., et al. (2013). Splicing proofreading at 5′ splice sites by ATPase Prp28p. Nucleic Acids Res. 41, 4660–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G.W., Coufal N.G., Liang T.Y., et al. (2009). An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16, 130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai B., Villen J., Beausoleil S.A., et al. (2008). Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7, 1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., and Darnell R.B. (2011). Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat. Biotechnol. 29, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.