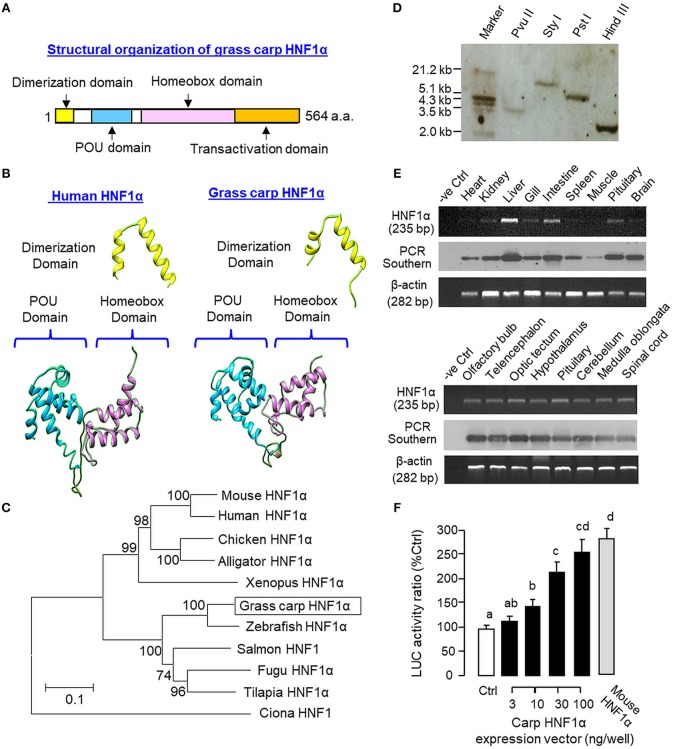
Figure 1.
Structural characterization, gene copy number, tissue distribution, and functional validation of grass carp HNF1α. (A) Structural organization of carp HNF1α a.a. sequence into four signature motifs, including the dimerization domain, POU domain, homeobox domain, and transactivation domain. (B) 3D Protein modeling of the dimerization domain, POU domain, and homeobox domain of carp HNF1α using the corresponding structures of their human counterparts as the template with SWISS-MODEL program. (C) Phylogenetic analysis of carp HNF1α cDNA sequence using the neighbor-joining method with MEGA 6.0 with the corresponding sequences identified in other species. The scale bar represents the evolutionary distance and the values for individual nodes of the guide tree are the percentage based on 1,000 bootstraps. (D) Copy number of HNF1α gene in the carp genome. Southern blot was conducted in genomic DNA after digestion with Pvu II, Sty I, Pst I, and Hind III, respectively, using a DIG-labeled cDNA probe for HNF1α. (E) Tissue expression profiling of HNF1α with RT-PCR. Total RNA was isolated from selected tissues and brain areas and subjected to RT-PCR using primers specific for carp HNF1α. The authenticity of PCR products was confirmed by PCR Southern and RT-PCR for β actin was used as the internal control. (F) Functional expression of carp HNF1α in αT3 cells. The ORF of grass carp HNF1α was subcloned into the eukaryotic expression vector pcDNA3.1 and transfected into αT3 cells with the Luc reporter pGL3.HNF1α.Luc with tandem repeats of HBE sites in its 5′promoter. In this study, parallel transfection with the expression vector for mouse HNF1α was used as a positive control. Data presented are expressed as mean ± SEM and groups denoted by different letters represent a significant difference at p < 0.05 (ANOVA followed by Newman-Keuls test).