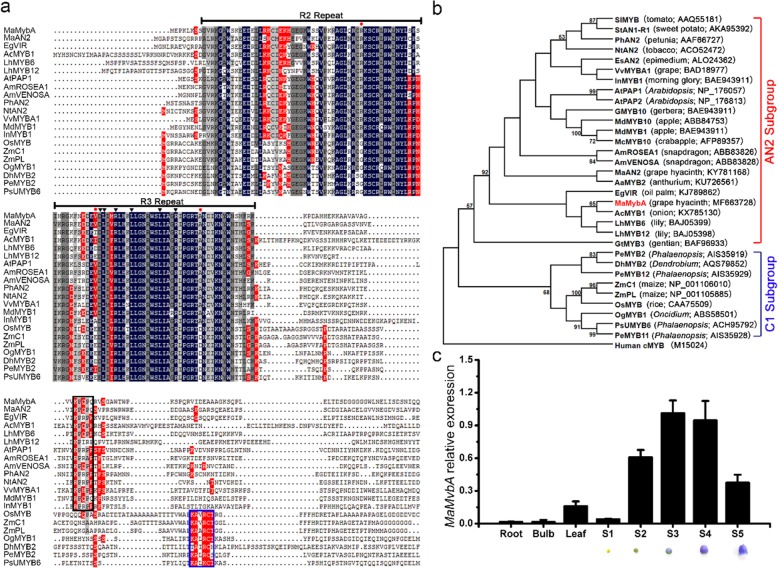
Fig. 1.
Multiple alignment, phylogenetic analysis, and transcription expression of MaMybA. a The sequence alignment of partial amino acids deduced from MaMybA, MaAN2, and other R2R3-MYBs. R2R3 repeats are indicated above the alignment. Three conserved amino acids [arginine (R), valine (V), and alanine (A)] in the R2R3 repeats of anthocyanin-promoting MYBs in dicots are marked using a red solid circle. The motif [D/E]LX2[K/R]X3LX6LX3R in the R3 repeat is necessary for interactions with a bHLH (basic Helix-Loop-Helix) protein, and is indicated by dark solid triangles. The typical motif [K/R]P[Q/R]P[Q/R] in the AN2 subgroup and KAX[K/R]C[S/T] in the C1 subgroup are shown by a black box and a blue box, respectively. b Phylogenetic analysis of entire amino acid sequences deduced from MaMybA, MaAN2, and other anthocyanin-related R2R3-MYBs. The phylogenetic tree was constructed by the maximum-likelihood method using MEGA 6.0 software. The numbers next to the nodes indicate bootstrap values from 1000 replicates. Human cMYB (M15024) was used as a rooting outlier. R2R3-MYB protein sequences of different plant species were retrieved from the GenBank database, and their GenBank accession numbers are listed in brackets. c Expression profile of MaMybA in different tissues of Muscari armeniacum; MaActin was the reference gene. Error bars indicate the standard deviations (SD) of average results