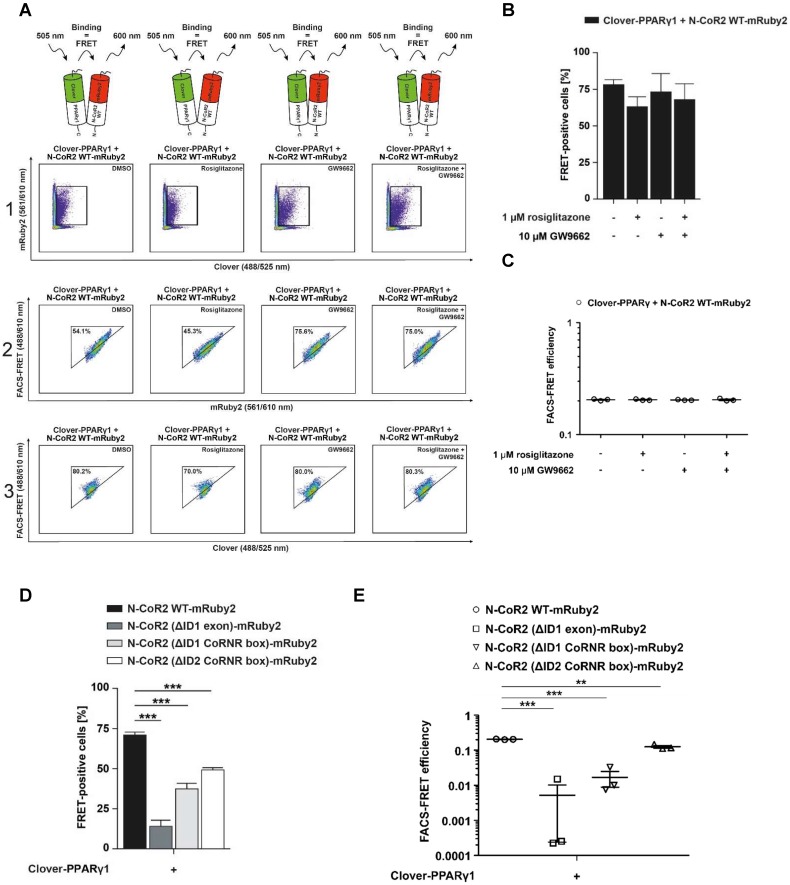
Figure 6.
Flow cytometry-based FRET-measurements to analyze protein-protein interactions between Clover-PPARγ1 and N-CoR2-mRuby2 constructs. Representative primary flow cytometry-plots showing the quantity of FRET-positive living HEK293T Clover-PPARγ1 + N-CoR2 WT-mRuby2 cells are depicted (A). Images are representative of experiments which were performed three times. The cells were cultured for 24 h with DMSO, 1 µM of the PPARγ agonist, rosiglitazone, and 10 µM of the PPARγ antagonist, GW9662, alone or in combination. Panel 1 (1) represents the gating for Clover/mRuby2-double positive cells. Panel 2 (2) depicts cells positive for the first FRET gate of FRET 488/610nm vs. mRuby2 561/610 nm and panel 3 (3) shows cells with a FRET signal. Numbers in panels (2) and (3) represent total percentages of cells within the FRET gates. A comparison of the total percentages of FRET-positive living HEK293T Clover-PPARγ1 + N-CoR2 WT-mRuby2 cells is depicted in (B). The relative flow cytometry-based FRET efficiencies calculated as described in Materials and Methods are presented in (C). Comparison of the total percentages of FRET-positive living HEK293T Clover-PPARγ1 + N-CoR2 WT-mRuby2, Clover-PPARγ1 + N-CoR2 (ΔID1 exon)-mRuby2, Clover-PPARγ1 + N-CoR2 (ΔID1 CoRNR box)-mRuby2 and Clover-PPARγ1 + N-CoR2 (ΔID2 CoRNR box)-mRuby2 cells are depicted in (D). Flow cytometry-based efficiencies of Clover/mRuby2-double positive cells, calculated as described in Materials and Methods, are shown in (E). Values are means ± SD of three experiments. **P ≤ 0.01 and ***P ≤ 0.001.