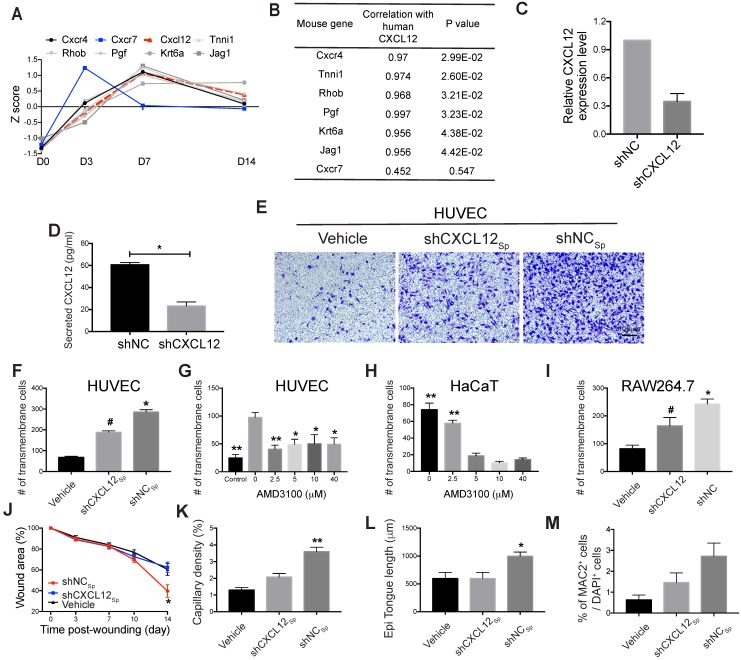
Figure 5.
Role of the CXCL12/Cxcr4 axis in EMSCSp-promoted wound healing. (A-B) Pearson's correlation coefficient between the expression of CXCL12 in EMSCSp and the expression of Cxcl12-targeted genes in mouse cells based on RNA-seq data from wounds treated with EMSCSp and vehicle 0, 3, 7, and 14 days after wound formation (A). The X-axis represents the time, and the Y-axis represents the z-score values of the coefficient. The correlations between Cxcl12-targeted genes with CXCL12 expression and their corresponding P values are listed in (B). (C) CXCL12 expression was decreased in EMSC stably transduced with lentivirus expressing shCXCL12 compared to that in EMSC transduced with lentivirus expressing shLacZ. The knockdown efficiency was determined by qPCR with 3 biological repeats. (D) CXCL12 secretion by EMSC transduced with shNC (control) or shCXCL12 was measured via ELISA. *P < 0.05 per the Mann-Whitney U test. (E-F) The migration of HUVECs towards EMSCSp expressing shCXCL12 was reduced compared to HUVEC migration towards EMSCSp expressing shNC in a Transwell assay. HUVECs were seeded on the top of the membrane insert, and no cells (vehicle, left), EMSCSp expressing shNC (shNCSp, right) or shCXCL12 (shCXCL12Sp, middle) were seeded on the bottom of the Transwell insert. HUVECs that migrated across the membrane were stained with crystal violet and photographed under a microscope. The numbers of migrated HUVECs in the three groups are displayed in a bar chart and represent 3 biological repeats. *P < 0.05 for shNCSp versus shCXCL12Sp or vehicle; and #P < 0.05 for shCXCL12Sp versus vehicle per ANOVA followed by Tukey's test. (G) Transwell assay to detect the EMSCSp-oriented migration of HUVECs pretreated with AMD3100 (a CXCR4 antagonist) at various concentrations for 30 min. EMSCSp were seeded on the bottom of the Transwell insert to induce HUVEC migration. After incubation for 24 h, the number of HUVECs that migrated across the membrane was counted. The results were determined in three independent experiments. *P < 0.05 and **P < 0.01 versus 0 μM AMD3100 per ANOVA followed by Tukey's test. (H) Transwell assay to detect the EMSCSp-oriented migration of HaCaT cells pretreated with AMD3100, with the number of cells that crossed the membrane counted as described above. The results were determined in three independent experiments. **P < 0.01 versus 5, 10 or 40 μM AMD3100 per ANOVA followed by Tukey's test. (I) Transwell assay to detect the EMSCSp-oriented migration of RAW264.7 macrophages. The number of cells that crossed the membrane was counted as described above. The results were determined in three independent experiments. *P < 0.05 for shNC versus shCXCL12 or vehicle and #P < 0.05 for shCXCL12 versus vehicle per ANOVA followed by Tukey's test. (J-M) Measurement of the wound area (J), capillary density (number of CD31+ cells among the total cells per wound section) (K), epithelial tongue length (L), and macrophage (MAC2+) percentage (M) in mice after transplantation with shNCSp, shCXCL12Sp or vehicle control. *P < 0.05 and **P < 0.01 for shNCSp versus shCXCL12Sp or vehicle per one-way ANOVA followed by Tukey's or Dunn's posttest.