Figure 3.
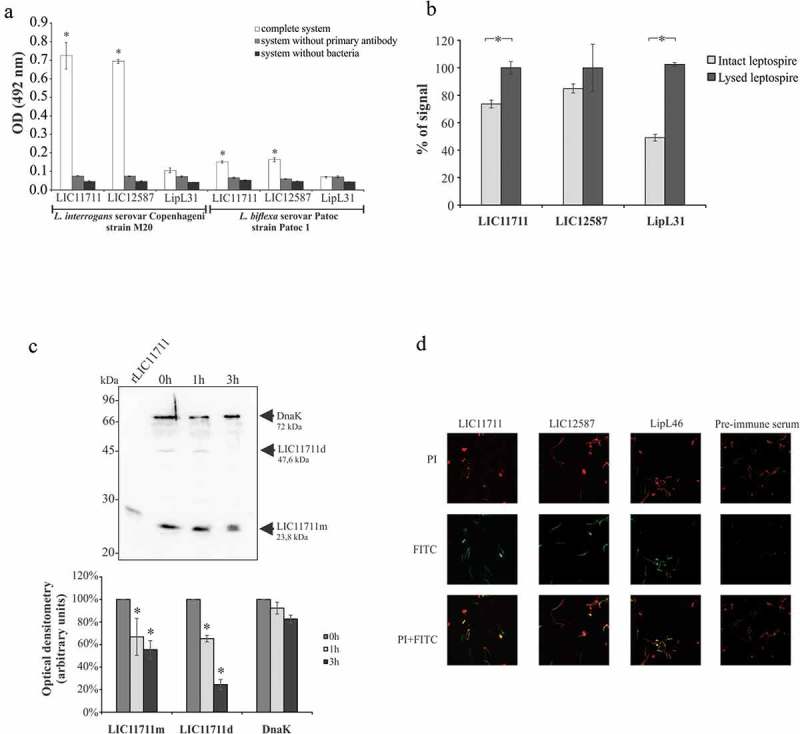
Cellular localization of native proteins in L. interrogans serovar Copenhageni. Evaluation of native proteins in intact (a) or lysed (b) L. interrogans serovar Copenhageni strain M20 by ELISA. Leptospires were immobilized on 96-well plates and incubated with polyclonal antiserum against recombinant protein followed by HRP-conjugated anti-mouse antibody addiction. L. biflexa was used as a control to differentiate pathogenic leptospires from non-pathogenic (a) and the cytoplasmic protein LipL31 was used as a parameter to differentiate surface exposed from cytoplasmic proteins. Statistical analyses were calculated comparing complete system with LipL31 in each group by Student’s t-test (*p < 0.05) (a). In (b) OD492nm values obtained in each group for the lysate treatment were considered 100% of signal and was compared to the signal obtained with the intact bacteria by Student’s t-test (*p < 0.05). (c) Protein accessibility by proteinase K degradation. Viable L. interrogans serovar Copenhageni (strain M20) cells were incubated with 25 μg/ml PK for 1 and 3 h. Cells were heated and cellular extracts were transferred to nitrocellulose membrane. For protein detection, membrane was incubated with polyclonal antiserum against rLIC11711 or rLIC12587 and DnaK (experimental control) followed by HRP-conjugated anti-mouse antibody (upper panel). Protein degradation after 1 and 3 h incubation with PK was measured by band densitometry and statistical analysis were calculated comparing with non-treated sample (0 h) by Student’s t-test (*p < 0.05). Representative results refer to one independent experiment out of two. m: monomer; d: dimer. (d) In immunofluorescence assay, L. interrogans serovar Copenhageni strain M20 were fixed and treated with the antiserum against rLIC11711, rLIC12587, LipL46 (positive control) or pre-immune serum (negative control). Leptospiral DNA was labeled by propidium iodide (PI). The co-localization was performed by overlapping the images of leptospires marked by PI and the target proteins labeled by fluorescein isothiocyanate (FITC).
