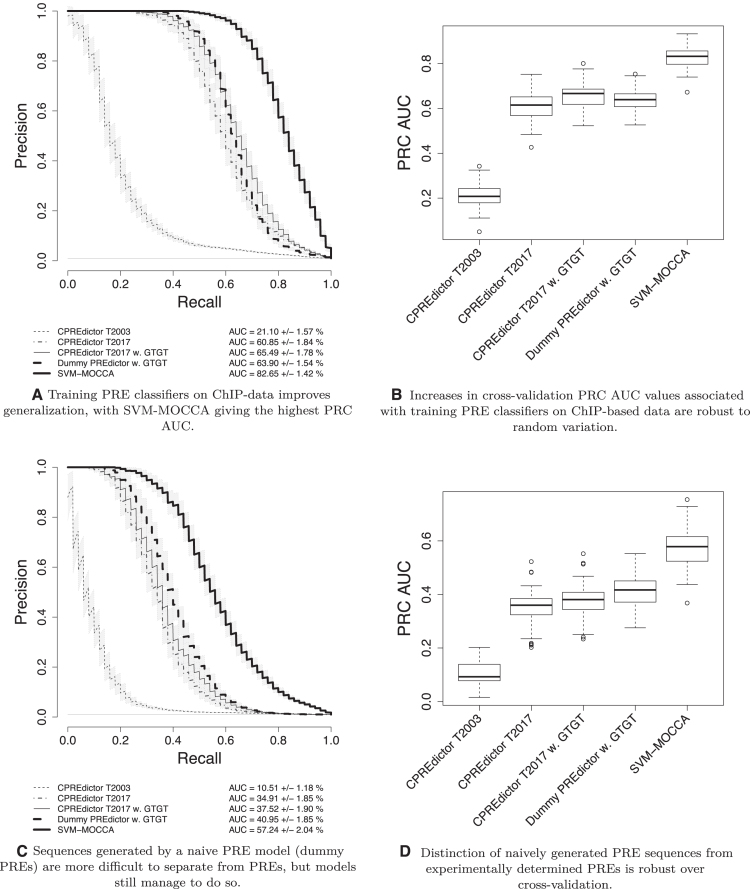
Figure 1.
Classifier generalization when trained on genome-wide experimental data for PRE prediction. (A) Average Precision/Recall plot for classifiers applied to PREs determined by Schwartz et al. (34) (independent from training set PREs) versus 100 times as many control sequences generated by a fourth-order Markov Chain trained genome-wide, as according to the plot legend. Average curves over all 50 folds are shown, together with 95% confidence intervals for the mean precision. AUC values are percentages rounded to two digits. (B) PRC AUC box plot for multiple classifiers over all 50 folds. (C) Average Precision/Recall plot for PREs determined by Schwartz et al. (34) (independent from training set PREs) versus 100 times as many sequences generated randomly using a fourth-order Markov Chain trained on PREs, constituting a naive PRE model (dummy PREs). Average curves over all 50 folds are shown, together with 95% confidence intervals for the mean precision. (D) PRC AUC box plot for multiple classifiers over all 50 folds.