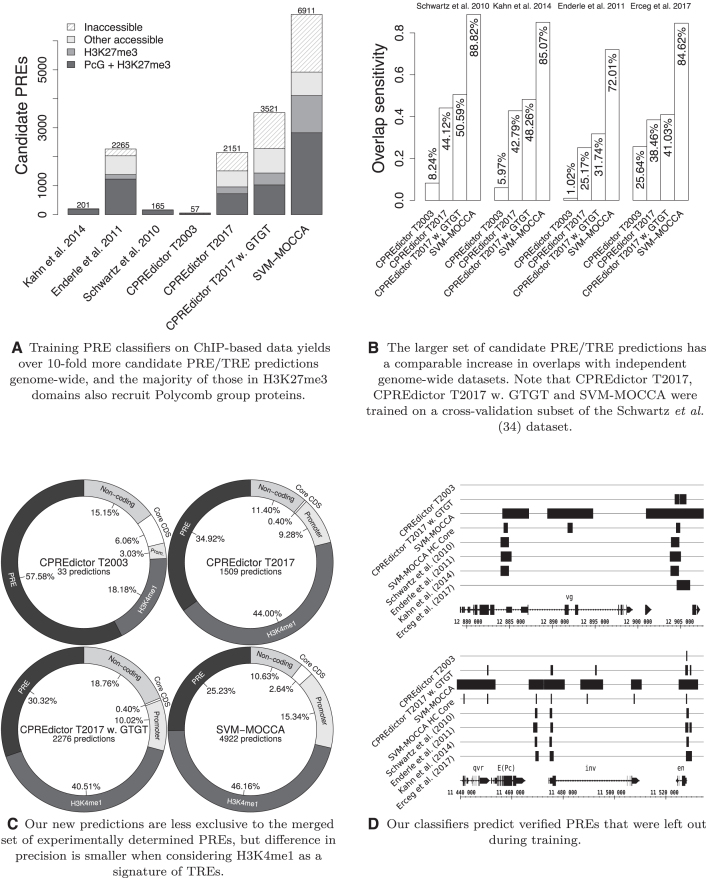
Figure 2.
Results of genome-wide candidate PRE/TRE prediction for an expected precision of 80%. (A) Numbers of experimentally determined and computationally predicted candidate PREs. Accessible portions in Polycomb repressed domains (H3K27me3) have been marked, as well as the portions of those regions that are enriched in Polycomb. Chromatin accessibility was derived from DNaseI-seq data; see Materials and Methods, also for H3K27me3 and Polycomb datasets. (B) Overlap sensitivity of each classifier’s predictions to two genome-wide, experimentally determined candidate PRE sets (35,36) and a set of functionally validated PREs (69) (see Materials and Methods for the definition of these three sets). Overlap sensitivity is defined as the fraction of regions in an experimental set that are overlapped by at least one prediction. (C) Proportions of the sets of predictions that overlap with different genomic loci. Only predictions in accessible chromatin are considered. The merged set of experimentally determined PREs by Kahn et al. (36), Enderle et al. (35) and Schwartz et al. (34) are considered first, and from the leftover, H3K4me1, then promoters, then core CDS; the final leftover set of predictions is marked as non-coding. See Materials and Methods for H3K27me3 datasets. Promoters are predicted as 3 kb upstream to 0.5 kb downstream from annotated gene transcription start sites. Core CDS is annotated coding sequence (CDS) shrunk bi-directionally by 250 bp (see Materials and Methods). (D) invected/engrailed and vestigial loci, visualized with the Integrated Genome Browser (63).