Fig. 2.
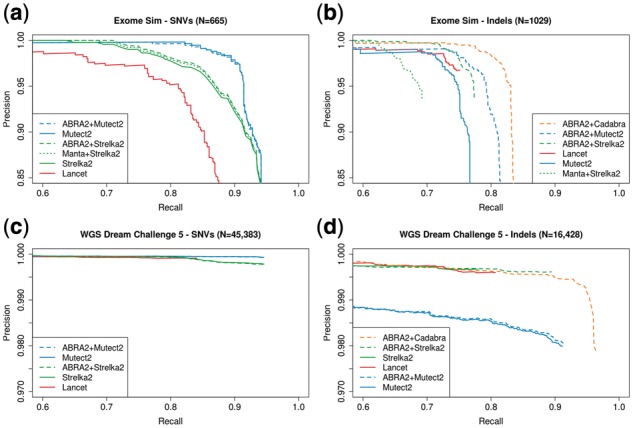
Evaluation of somatic variant calling in DNA. (a) Precision and recall of somatic SNV detection on a challenging simulated exome dataset containing insertions and deletions ranging in length from 1 to 100 bp and variant allele frequency ranging from 10 to 50%. Strelka2, Strelka2/Manta, Mutect2 and Lancet are evaluated with Strelka2 and Mutect2 also applied to ABRA2 realignments. (b) Precision and recall of somatic indel detection on the exome dataset. In addition to the SNV callers, Cadabra is applied to the ABRA2 realignments. Pipelines including ABRA2 produce the best overall results. (c) Somatic SNV calling on the ICGC Dream Somatic Mutation Calling Challenge 5 dataset. Strelka2, Mutect2 and Lancet are evaluated with Strelka2 and Mutect2 also applied to ABRA2 realignments. (d) Somatic indel calling on the ICGC Dream Somatic Mutation Calling Challenge 5 dataset. In addition to the SNV callers, Cadabra is applied to the ABRA2 realignments. ABRA2 combined with Cadabra or Strelka2 produces the best overall results
