Fig. 6.
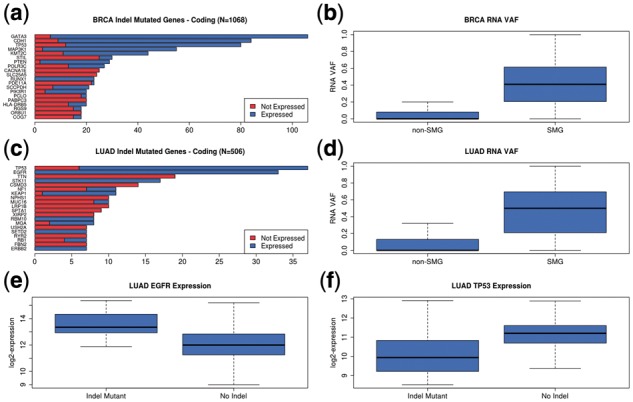
TCGA breast and lung adenocarcinoma indels. (a) Frequency of genes harboring somatic coding indels in the TCGA BRCA cohort with evidence of expression of the indel mutation. Frequency is computed by simple counts of subjects containing coding mutations and does not take into account exon lengths of each gene. (b) Comparison of RNA Variant Allele Frequency (VAF) of coding indels between genes previously identified as ‘significantly mutated’ (SMG) and not significantly mutated (non-SMG) in the BRCA cohort. (c) Frequency of genes harboring somatic coding indels in the TCGA LUAD cohort with evidence of expression of the indel mutation. (d) Comparison of RNA VAF of coding indels between genes previously identified as ‘significantly mutated’ and not significantly mutated in the LUAD cohort. (e) EGFR gene expression for tumor RNA samples containing an EGFR coding indel versus those without an EGFR coding indel in the TCGA LUAD cohort. EGFR is more highly expressed in samples containing an indel. (f) TP53 gene expression for tumor RNA samples containing a TP53 coding indel versus those without a TP53 indel in the TCGA LUAD cohort. TP53 expression is lower in samples containing an indel
