Abstract
Residual energy intake (REI) on two successive diets (hay and maize based) and slaughter traits, including visceral organs, were phenotyped in 584 adult purebred Charolais cows. To investigate the relationships between these traits and their genetic determinism, we first estimated the genetic parameters, including correlations, using REML modeling under WOMBAT software. The animals were then genotyped on the BovineSNP50 SNPchip before being imputed to the 600K density and genome wide association study was performed with GCTA software. We found low heritability for REI (h2 = 0.12 in each of the diet phases). Although the phenotypic correlation between the two diet phases was moderate (0.36), the genetic correlation was high (0.83), indicating a common genetic determinism for feed efficiency regardless of the diet. Correlations between REI and slaughter traits were negative regarding muscle-related traits and positive for fat-related traits, indicating that efficient animals generally had a more muscular carcass. It was also seen that feed efficiency was genetically and phenotypically correlated with smaller organs when expressed as a proportion of their empty body weight. From the GWAS analysis, seven QTLs were found to be associated with a trait at the genome-wide level of significance and 18 others at the chromosome-wide level. One important QTL was detected in BTA 2, reflecting the essential effect of the myostatin gene on both carcass composition and relative organ weight. Three QTLs were detected for REI during the maize diet phase on BTA 13, 19, and 28, the latter being significant at the genome-wide level. The QTLs on BTA 19 mapped into the TANC2 gene and the QTLs on BTA 28 into the KIF1BP gene, which are both known to interact with the same protein (KIF1A). However, no obvious functional link between these genes and feed efficiency could be made. Among the other QTLs detected, one association on BTA 4 with liver proportion mapped to the candidate gene WASL, which has previously been shown to be differentially expressed in liver cells and linked to feed restriction or cancer development. No QTLs were found to be common between feed efficiency and any slaughter traits.
Keywords: beef cattle, feed efficiency, genetic correlations, GWAS, slaughter traits
INTRODUCTION
In the current context of high feed costs and competition for food between livestock and humans, the issue of feed efficiency has become critical in cattle production. Improving the balance between output (production) and input (feed intake) is central to ensuring the economic and environmental sustainability of the beef and dairy industries. The traditional method to measure feed efficiency is to use a ratio trait such as the feed conversion ratio (i.e., the ratio between feed intake and body weight gain) or its reverse, gain efficiency. However, genetic selection on a ratio trait is generally not considered as optimal (Gunsett, 1984; Campo and Rodríguez, 1990). Indeed, this type of selection may only affect one of the two parts of the ratio, and this part may not be constant over animals and time, resulting in a lack of selection efficiency. Despite a recent study showing some efficiency of selection on a ratio trait in pigs (Shirali et al., 2018), this method is not widely used in cattle. Another means of determining feed efficiency is to study residual feed intake (RFI). First proposed by Koch et al. (1963), RFI is the difference between the actual feed intake of an animal and its predicted feed intake based on its performance (i.e., the intake necessary to cover the demands of different energy sinks) estimated by regression. Because of its residual nature, RFI includes both modeling errors and the actual variability in efficiency. However, this definition allows RFI to be conceptually phenotypically independent of the traits used for prediction and, therefore, reflects digestive and metabolic variabilities (Archer et al., 1999; Herd et al., 2004; Berry and Crowley, 2013). For this reason, RFI has become the measure of choice for feed efficiency in cattle (Kenny et al., 2018).
Feed efficiency is a complex and multifactorial trait and the biological processes involved change throughout the life of an animal. Energy sinks are indeed dependent on the context of the animal: growth, gestation, lactation, accretion, or mobilization of body reserves, physical activity, etc. Moreover, an animal also needs to maintain its integrity and deal with the protein turnover that contributes to this expenditure. This maintenance requirement can be defined as the energy required by an animal to sustain a perfectly stable body weight (Archer et al., 1999). Studies have suggested that maintenance requirements represent the principal energy sink, using up to 75% of the total energy requirement in some situations, but considerable variabilities exist between animals (Klosterman, 1972; Ferrell and Jenkins, 1985; Montaño-Bermudez et al., 1990; Richardson and Herd, 2004; Kenny et al., 2018). This variability may have a metabolic source or result from differences in resource allocation between energy sinks. Numerous studies have indeed reported an association between maintenance requirements and body composition, as well as possible links between these requirements and the relative size of visceral tissues and organs (reviewed by Archer et al., 1999 and Kenny et al., 2018). Digestive efficiency is also a component of feed efficiency. Individual variations in digestive abilities have been demonstrated and may be associated with the microbiome (Khiaosa-ard and Zebeli, 2014). Digestibility is also strongly dependent on the type of feed offered to the animal.
Studies have already started to explore the genomic control of feed efficiency, and quantitative trait locus (QTL) regions have been evidenced in growing animals (e.g., Nkrumah et al., 2007b; Rolf et al., 2012; Lu et al., 2013; Seabury et al., 2017). In this study, the aim was to explore feed efficiency in adult cows during the preslaughter period, which represent a new type of animal in this type of studies. This enabled us to explore feed efficiency when maintenance was almost the only energy sink. Based on a design that allowed individual data acquisition under two successive diets and detailed carcass traits at slaughter, we studied, at both the phenotypic and genetic levels, the influence of roughage type on feed efficiency, as well as its links to body composition and visceral organ weight. Association studies were performed on efficiency traits and on carcass traits, some of which are rarely phenotyped.
MATERIALS AND METHODS
Ethics Statement
During this experiment, all animals were kept indoors and handled with care in line with Institut National de la Recherche Agronomique’s (INRA) ethics policy in compliance with the guidelines for animal research issued by the French Ministry of Agriculture (https://www.legifrance.gouv.fr/eli/decret/2013/2/1/2013–118/jo/texte). All blood samples were drawn by the appropriate staff on the farm who had been trained by veterinarians.
Animals and Feed Management
The experiment was performed at the Bourges-La Sapinière Experimental Farm (France) belonging to the INRA between 1985 and 2010. A total of 584 purebred Charolais females were procreated from 60 insemination sires. After a first calving at 3 yr of age and three further calvings and lactations, dry females were managed during a preslaughter period, which is the subject of the current study. This preslaughter period comprised an adaptation period of 4 wk with a hay diet (not studied here), a first-studied phase of 4 wk when they received a hay-only diet ad libitum, followed immediately by a second-studied phase involving a maize silage-based diet. The latter diet contained ad libitum maize silage plus 1,800 g soy meal, 0.60 g urea, and 300 g minerals per head daily. The maize phase lasted on average 6 wk and ended when the farm manager took the decision to slaughter the animal as a function of its body condition. The animals were accommodated in pens (seven per pen) equipped with individual troughs and automatic gates (American Calan Inc., Northwood, NH, USA) so that their individual feed intake could be measured.
Fattening and Feed Efficiency Traits
Individual feed intake was determined by weighing the forage distributed every day and the forage refused three times a week. Both the forage distributed and that refused were sampled three times a week for analysis. The hay dry matter (DM) was on average 84.2% (SD 2.4) with an energy value of 0.57 Unité Fouragère Viande (UFV; SD 0.04) per kg DM. The maize silage DM was on average 35.5% (SD 3.4) with an energy value of 0.85 UFV (SD 0.03) per kg DM. The animals were weighed on two consecutive days at the start of the hay phase, at transition from hay to maize and at the end of the maize phase. They were also weighed every 2 wk during these phases. Their body condition score (BCS) was evaluated at the start and end of each phase, as well as at all intermediate points.
Because the phases were not of the same duration in all animals, intra-animal regressions were performed on the body weight and BCS using the general linear model (GLM) procedure under SAS/STAT software, version 9.4 of the SAS System for Linux (Copyright 2002 to 2012 by SAS Institute Inc., Cary, NC, USA). This made it possible to define initial, mid-test, and final values for body weight and BCS for each phase and on a common timeline for all animals. The duration of the hay phase was fixed at 28 d and that of the maize phase at 48 d. The initial and final weights were used to compute the average daily gain (ADG), defined as the coefficient of the linear regression of body weight over time, and the mean metabolic weight (MMW), calculated as (0.5[end weight of phase + start weight of phase])0.75.
Because of these two diets, we decided to express feed efficiency in terms of energy rather than kilogram of feed. The residual energy intake (REI) was therefore used rather than the RFI. REI was defined by the difference between observed and expected energy intake, computed by a regression of energy intake on MMW and ADG using the GLM procedure under SAS/STAT software according to the following model:
REIs were calculated separately for each phase. According to this model, efficient animals were those with negative REI values.
Means and SDs were calculated for each trait using the Proc Means under SAS/STAT software. Animals with performance deviating by more than 4 SD from the mean were discarded for the analyses, so ultimately 578 females from 59 different sires were studied. The average family size was 10.3 females (SD 6.9) ranging from 2 to 31.
Slaughter Traits
Animals were slaughtered at INRA’s experimental slaughterhouse in Theix (France). The weight of hot carcasses was recorded. After chilling for 24 h, the sixth rib was excised and dissected in order to estimate the carcass muscle and fat contents. Of the visceral organs, the fifth quarter fat, rumen, omasum, abomasum, intestines, liver, lungs, heart, kidneys, and spleen were weighed, together with the reproductive tract. Hot carcass and organ weights were expressed relative to the empty body weight (EBW). Muscle and fat weights in the carcass were estimated using following predictive equations based on those developed by Robelin and Geay (1975):
The carcass muscle and fat contents were calculated by dividing the estimated muscle and fat weights by the hot carcass weight.
Statistical Analysis
Genetic parameters.
Genetic parameters were estimated using the restricted estimation of maximum likelihood (REML) method under WOMBAT software (Meyer, 2007). The correlation between the hay REI and maize REI was estimated using a bivariate linear animal model, while correlations with other variables were estimated using a trivariate linear animal model, including the two REI and the given variable. The model considered for all traits can be expressed in a matrix notation as:
where y is the vector of observations for the trait, b is the vector containing the year effect (considered as a fixed effect), a is the vector of animal additive genetic effects, e is the vector of residuals and X and Z are the respective incidence matrices assigning observations to effects. Random effects were assumed to be normally distributed with means equal to 0 and a covariance structure equal to
where G is a (co)variance matrix of random direct additive genetic effects and R is the residual (co)variance matrix. The A matrix represents the additive genetic relationships between animals, and Ir is the identity matrix which has an order equal to the levels of appropriate residuals effect.
The pedigree file consisted of five generations that included 2,023 animals. Trait heritability was estimated from the ratio between the animal variance component and the sum of the animal variance component and the residual variance.
Genome-wide association studies
Animals were genotyped using the BovineSNP50 (50K) SNPchip (Illumina Inc., San Diego, CA, USA). DNA extraction from blood samples and genotyping were performed at the Laboratoire d’Analyses Génétiques pour les Espèces Animales in Jouy-en-Josas, France (LABOGENA; www.labogena.fr). Using FImpute software (Sargolzaei et al., 2014), all genotypes were imputed to the high-density SNPchip. The reference population consisted of 664 French Charolais animals, with an allelic imputation error rate lower than 1% (Hozé et al., 2013). The marker order and positions were based on the UMD3.1 bovine assembly. A total of 647,179 autosomal single nucleotide polymorphisms (SNPs) and 496 animals were retained after applying the quality control filters used in the French national evaluation system (Boichard et al., 2012): an individual call rate higher than 95%, an SNP call rate higher than 90%, a minor allele frequency higher than 5% in at least one major French dairy cattle breed, and genotype frequencies in Hardy–Weinberg equilibrium with P > 10−4. Stratification of the population was assessed by principal component analysis (PCA) computed with plink software (Purcell et al., 2007). The association analyses were single trait and performed using the mlma option of GCTA software (Yang et al., 2011); this applies a mixed linear model that includes the candidate variant:
where y is the vector of phenotypes (corrected for the year effect using the covar option); µ is the overall mean; b is the additive fixed effect of the candidate variant to be tested for association; x is the vector of predicted allele assays ranging from 0 to 2; u ~ N(0, G) is the vector of random polygenic effects (with G being the genomic relationship matrix, calculated using high-density SNP genotypes, and being the estimated polygenic variance based on the null model [y = μ + u +e] and then fixed while testing for the association between each variant and the trait) and e ~ N(0, I) is the vector of random residual effects, with I being the identity matrix and the residual variance. Because of the test multiplicity, a Bonferroni correction of α = 5% was applied to both genome-wide and chromosome-wide thresholds (threshold of significance = −log10[α/number of SNPs]). SNPs with P < 7.72 × 10−8 were considered to be significantly associated at the genome-wide level. The chromosome-wide thresholds ranged from P < 4.23 × 10−6 to P < 1.24 × 10−6. The confidence interval of the QTL location was estimated using the logarithm drop-off method in order to include SNP with a probability higher than 1/1000 of SNPmax probability.
Significant regions were compared with already identified QTLs recorded in the cattle QTL database https://www.animalgenome.org/cgi-bin/QTLdb/index. In addition, biological functions of strong positional candidate genes were reviewed using https://www.uniprot.org/ in order to identify possible functional candidates among them.
RESULTS
Descriptive Statistics
Descriptive statistics are reported in Table 1 for feed efficiency traits and Table 2 for slaughter traits. Although the average raw feed consumption (expressed in kilogram per day) was almost three time higher during the maize phase than the hay phase, this difference disappeared when intakes were expressed in kilogram DM. Obviously, maize silage contains more energy than hay (per kilogram DM), so the energy intake was on average 40% higher during the maize phase. The average energy intake during the hay phase (6.82) was slightly higher than the estimated 6.3 UFV/d that would have been necessary to meet maintenance requirements alone, according to INRA equation (INRA, 2018). This was in line with the observations as the animals were able to gain a small amount of weight (0.49 kg/d) and body condition (0.18) during the period. During the maize phase and because the females were heavier, their maintenance requirements were slightly higher and estimated at an average of 6.6 UFV/d. During this second phase, the actual energy intake was substantially higher at 9.53 UFV/d. The increases in body weight and condition were consequently greater: 1.18 kg/d and 0.68, respectively. These gains in weight and body condition were expected and appropriate during this experiment where the animals were in their finishing and preslaughter period. By construction, the REI was an average of zero in each phase. For the hay phase, the R2 of the regression was 0.15, while it was 0.49 for the maize phase. The ADG did not have a significant effect on the regression for the hay phase.
Table 1.
Means, SDs, minimums (Min.), and maximums (Max.) of feed efficiency traits for both the hay and maize phases
| Hay phase | Maize phase | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | n | Mean | SD | Min. | Max. | Mean | SD | Min. | Max. |
| Feed intake, kg/d | 578 | 14.12 | 3.10 | 4.77 | 26.34 | 32.23 | 5.97 | 13.59 | 51.56 |
| DMI, kg/d | 578 | 11.96 | 2.65 | 4.07 | 21.99 | 11.21 | 2.12 | 4.89 | 18.13 |
| Energy intake, UFV/d | 578 | 6.82 | 1.51 | 2.32 | 12.53 | 9.53 | 1.80 | 4.16 | 15.4 |
| Initial weight, kg | 578 | 707.85 | 70.39 | 479.45 | 938.79 | 719.83 | 70.93 | 482.22 | 927.87 |
| Final weight, kg | 578 | 721.56 | 70.55 | 479.17 | 933.59 | 769.27 | 74.77 | 528.82 | 1,003.27 |
| Average daily gain, kg/d | 578 | 0.49 | 0.70 | −1.51 | 2.44 | 1.18 | 0.53 | −0.70 | 2.80 |
| Initial BCS | 578 | 2.39 | 0.51 | 1.00 | 5.00 | 2.55 | 0.50 | 1.00 | 5.00 |
| Final BCS | 578 | 2.57 | 0.50 | 1.00 | 5.00 | 3.23 | 0.60 | 1.00 | 5.00 |
| REI, UFV | 578 | 0.00 | 1.39 | −4.35 | 5.46 | 0.00 | 1.28 | −5.87 | 4.11 |
Table 2.
Means, SDs, minimums (Min.), and maximums (Max.) for slaughter traits
| Trait | Number of animals | Mean | SD | Min. | Max. |
|---|---|---|---|---|---|
| EBW, kg | 542 | 634.45 | 58.88 | 433.06 | 853.35 |
| Carcass, % of EBW | 542 | 65.79 | 1.64 | 59.87 | 71.96 |
| Muscle, % of carcass | 542 | 64.91 | 3.28 | 54.66 | 76.34 |
| Fat, % of carcass | 542 | 19.42 | 3.14 | 8.81 | 30.27 |
| Fifth quarter, % of EBW | 519 | 33.22 | 1.68 | 25.64 | 37.40 |
| Leather, % of EBW | 520 | 7.74 | 0.73 | 5.62 | 9.88 |
| Fifth quarter fat, % of EBW | 542 | 4.12 | 0.90 | 1.33 | 7.80 |
| Rumen, % of EBW | 542 | 2.37 | 0.25 | 0.30 | 3.05 |
| Omasum, % of EBW | 542 | 1.01 | 0.22 | 0.34 | 1.97 |
| Abomasum, % of EBW | 542 | 0.45 | 0.08 | 0.27 | 0.87 |
| Intestines, % of EBW | 542 | 2.32 | 0.37 | 0.04 | 3.41 |
| Digestive tract, % of EBW | 542 | 6.15 | 0.62 | 3.17 | 8.32 |
| Reproductive tract, % of EBW | 493 | 0.37 | 0.14 | 0.11 | 0.84 |
| Liver, % of EBW | 542 | 1.17 | 0.11 | 0.82 | 1.70 |
| Lungs, % of EBW | 520 | 0.74 | 0.09 | 0.51 | 1.46 |
| Heart, % of EBW | 520 | 0.43 | 0.04 | 0.32 | 0.69 |
| Kidneys, % of EBW | 520 | 0.19 | 0.02 | 0.14 | 0.37 |
| Spleen, % of EBW | 519 | 0.17 | 0.02 | 0.11 | 0.24 |
Genetic Parameters
The heritability of REI in both the hay and maize phases and the correlations between these two traits are presented in Table 3. Heritability during the two phases was low and similar at around 0.12 (SE 0.08). Although the phenotypic correlation was already moderate (0.36), the genetic correlation was extremely high (0.83), suggesting that a considerable proportion of the genetic determinism underpinning feed efficiency might be shared between the two phases despite the difference in diet.
Table 3.
Heritability (on the diagonal), genetic correlations (above the diagonal), and phenotypic correlations (below the diagonal) for REI measured during the hay and maize phases, with SEs in brackets
| Trait | REI hay phase | REI maize phase |
|---|---|---|
| REI hay phase | 0.12 (0.08) | 0.83 (0.29) |
| REI maize phase | 0.36 (0.04) | 0.13 (0.08) |
The heritability of the other traits and their correlations with hay and maize REI are presented in Table 4, and they varied depending on the trait. The three weight traits (hay phase mid-weight, maize phase mid-weight, and EBW) are highly heritable, with a value of h2 = 0.68. By contrast, the heritability of components such as the lung or reproductive tract (expressed as a percentage of EBW) were quite low, at h2 = 0.05 and h2=0.08, respectively. Other slaughter traits were intermediate, with the heritability of digestive tract components ranging from 0.11 to 0.23 and that of other organs and carcass composition ranging from 0.27 to 0.53. For the ADG, heritability of the trait for the maize phase was slightly higher than that for the hay phase (0.13 and 0.08). The same pattern was observed with respect to energy intake (h2 = 0.15 for the hay phase and h2 = 0.20 for the maize phase). However, in light of the SEs, none of these differences between phases was significant.
Table 4.
Heritability (h2) and phenotypic (rp) and genetic (rg) correlations with REI measured during the hay and maize phases for various traits, with SEs in brackets
| Hay REI | Maize REI | ||||
|---|---|---|---|---|---|
| Trait | h 2 | r p | r g | r p | r g |
| Hay energy intake, UFV/d | 0.15 (0.08) | 0.97 (0.01) | 0.88 (0.07) | 0.36 (0.04) | 0.68 (0.28) |
| Maize energy intake, UFV/d | 0.20 (0.08) | 0.32 (0.04) | 0.60 (0.29) | 0.87 (0.01) | 0.68 (0.17) |
| Hay phase mid-weight, kg | 0.68 (0.08) | 0.01 (0.04) | 0.09 (0.23) | −0.01 (0.04) | 0.03 (0.23) |
| Maize phase mid-weight, kg | 0.68 (0.08) | 0.01 (0.04) | 0.11 (0.23) | 0.01 (0.04) | 0.04 (0.23) |
| Hay phase ADG, kg/d | 0.08 (0.07) | 0.00 (0.04) | 0.18 (0.54) | 0.10 (0.04) | −0.32 (0.58) |
| Maize phase ADG, kg/d | 0.13 (0.07) | 0.00 (0.04) | −0.14 (0.44) | 0.00 (0.04) | −0.17 (0.43) |
| EBW, kg | 0.68 (0.09) | 0.01 (0.04) | −0.12 (0.25) | 0.02 (0.02) | −0.12 (0.25) |
| Carcass, % of EBW | 0.53 (0.11) | −0.08 (0.05) | −0.24 (0.28) | −0.12 (0.05) | −0.32 (0.26) |
| Muscle, % of carcass | 0.33 (0.10) | −0.11 (0.04) | −0.44 (0.30) | −0.09 (0.04) | −0.35 (0.29) |
| Fat, % of carcass | 0.27 (0.09) | 0.11 (0.04) | 0.30 (0.33) | 0.09 (0.04) | 0.28 (0.31) |
| Fifth quarter, % of EBW | 0.51 (0.10) | 0.08 (0.05) | 0.62 (0.25) | 0.12 (0.05) | 0.43 (0.26) |
| Leather, % of EBW | 0.47 (0.11) | −0.03 (0.04) | 0.09 (0.30) | 0.01 (0.04) | 0.24 (0.28) |
| Fifth quarter fat, % of EBW | 0.33 (0.10) | 0.15 (0.04) | 0.59 (0.28) | 0.08 (0.04) | 0.25 (0.30) |
| Rumen, % of EBW | 0.23 (0.09) | 0.07 (0.04) | 0.49 (0.33) | 0.11 (0.04) | −0.11 (0.35) |
| Omasum, % of EBW | 0.19 (0.08) | 0.06 (0.04) | 0.12 (0.37) | 0.09 (0.04) | 0.15 (0.36) |
| Abomasum, % of EBW | 0.14 (0.08) | 0.04 (0.04) | 0.52 (0.40) | 0.09 (0.04) | 0.94 (0.40) |
| Intestines, % of EBW | 0.11 (0.07) | 0.09 (0.04) | 0.66 (0.47) | 0.11 (0.04) | 0.74 (0.41) |
| Digestive tract, % of EBW | 0.20 (0.08) | 0.11 (0.04) | 0.59 (0.35) | 0.16 (0.04) | 0.48 (0.34) |
| Reproductive tract, % of EBW | 0.08 (0.08) | −0.03 (0.04) | 0.83 (0.60) | 0.01 (0.04) | 0.72 (0.60) |
| Liver, % of EBW | 0.39 (0.09) | 0.11 (0.04) | 0.44 (0.27) | 0.26 (0.04) | 0.57 (0.26) |
| Lungs, % of EBW | 0.05 (0.07) | 0.06 (0.04) | 0.60 (0.68) | 0.06 (0.04) | 0.53 (0.66) |
| Heart, % of EBW | 0.50 (0.10) | −0.01 (0.05) | 0.34 (0.28) | 0.03 (0.05) | 0.25 (0.28) |
| Kidneys, % of EBW | 0.27 (0.09) | 0.01 (0.05) | 0.30 (0.35) | 0.07 (0.05) | 0.24 (0.34) |
| Spleen, % of EBW | 0.38 (0.10) | 0.02 (0.05) | 0.77 (0.26) | 0.03 (0.04) | 0.52 (0.31) |
The phenotypic correlations between REI and the corresponding mid-weight and ADG intraphase values were equal to zero, as had been anticipated in view of the definition of the REI equation. By contrast, correlations with energy intake were highly significant, even at the genetic level, with genetic correlations of between 0.60 and 0.88. Genetic correlations with mid-weights and the ADG were, however, mainly low and all were nonsignificant.
Phenotypic correlations with slaughter traits were low, ranging generally from −0.15 to 0.15, the only exceptions being the digestive tract (0.16) and liver (0.26) with the maize REI. For genetic correlations, the SEs were high (mostly around 0.3–0.4), thus limiting the number of significant results. The two REI measures tended to be negatively genetically correlated with the carcass muscle percentage (rg = −0.44 and rg = −0.35) and positively correlated with the carcass fat percentage (rg = 0.30 and rg = 0.28). This seems to indicate that efficient animals produced more carcass muscle than fat, although none of these correlations differed significantly from zero.
The importance of the fifth quarter and fifth quarter fat was highly positively correlated with the REI, particularly for the hay REI where the correlations were significant (rg = 0.62 and rg = 0.59). This indicates that efficient animals, particularly those on a hay-based diet, seemed also to be those with the fifth quarter and fifth quarter fat representing a smaller proportion of their EBW. This observation was confirmed by the fact that the genetic correlations between REI and organs were all positive (and with just one exception, nonsignificant). However, due to the large SE among all correlations between REI and organs, only three are significant: the correlation between maize REI and abomasum that was extremely high (rg = 0.94) and the correlations between maize REI and liver (rg = 0.57) and between hay REI and spleen (rg = 0.77).
Association Analyses
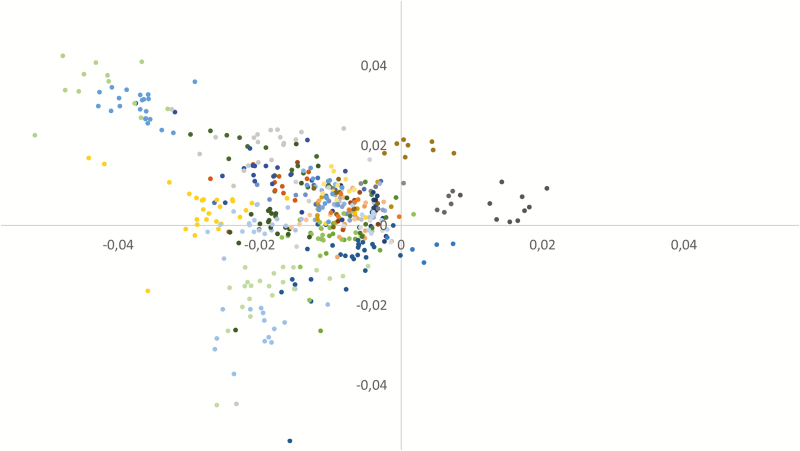
Stratification of the population, as evaluated by PCA, is shown in Figure 1. The population was generally homogenous but with a few families displaying slight differences. This structure was taken into account in the GWAS model using the genomic matrix.
Figure 1.
Population diversity shown following PCA (one color corresponds to one sire).
The results of GWAS analyses are presented in Table 5. Seven QTLs were found to be associated with a trait with a genome-wide level of significance, and 18 others were significant at the chromosome-wide level, thus giving a total of 25 QTLs scattered over 13 chromosomes and associated with 15 different traits. Overall, among the 26 different traits analyzed, no significant QTLs were found for 11 of them, one significant QTL was found for 8 of them, two QTLs were found for 4 of them, and three QTLs were found for 3 of them. The REI for the maize phase was the only nonslaughter trait for which QTLs were found.
Table 5.
List of all significant GWAS signals on the different traits
| BTA | Physical position (Mb) | Trait | Associated P-value | Number of significant SNP | Significant interval (Mb) | Level of significance1 |
|---|---|---|---|---|---|---|
| 1 | 110.27 | Omasum, % of EBW | 4.54 × 10−7 | 10 | 110.01–110.27 | ** |
| 2 | 6.22 | Intestines, % of EBW | 3.78 × 10−7 | 13 | 5.51–6.22 | ** |
| 2 | 6.22 | Carcass, % of EBW | 5.87 × 10−24 | 2 | 6.21–6.22 | *** |
| 2 | 6.22 | Digestive tract, % of EBW | 6.08 × 10−7 | 20 | 5.30–6.22 | ** |
| 2 | 6.22 | Fifth quarter fat, % of EBW | 1.00 × 10−10 | 5 | 5.59–6.22 | *** |
| 2 | 6.22 | Muscle, % of carcass | 4.74 × 10−26 | 2 | 6.21–6.22 | *** |
| 2 | 6.22 | Fat, % of carcass | 2.22 × 10−16 | 2 | 6.21–6.22 | *** |
| 2 | 6.22 | Leather, % of EBW | 6.51 × 10−7 | 12 | 5.49–6.22 | ** |
| 2 | 6.22 | Liver, % of EBW | 6.11 × 10−9 | 5 | 5.59–6.22 | *** |
| 2 | 6.22 | Fifth quarter, % of EBW | 5.44 × 10−29 | 2 | 6.21–6.22 | *** |
| 2 | 55.42 | Heart, % of EBW | 1.73 × 10−7 | 5 | 55.35–55.45 | ** |
| 2 | 76.43 | Rumen, % of EBW | 9.70 × 10−7 | 9 | 75.91–76.95 | ** |
| 3 | 30.59 | Abomasum, % of EBW | 1.28 × 10−6 | 5 | 30.59–30.71 | ** |
| 4 | 88.83 | Liver, % of EBW | 1.18 × 10−6 | 52 | 88.12–89.91 | ** |
| 7 | 70.95 | Lungs, % of EBW | 1.29 × 10−6 | 2 | 70.94–70.95 | ** |
| 8 | 74.83 | Leather, % of EBW | 4.77 × 10−7 | 47 | 72.49–75.15 | ** |
| 12 | 36.34 | Fifth quarter, % of EBW | 4.52 × 10−7 | 10 | 33.94–36.34 | ** |
| 13 | 9.256 | REI maize phase | 1.92 × 10−6 | 6 | 8.25–10.20 | ** |
| 13 | 27.01 | Lungs, % of EBW | 2.41 × 10−6 | 15 | 26.46–27.34 | ** |
| 19 | 48.17 | REI maize phase | 4.51 × 10−7 | 71 | 42.26–48.20 | ** |
| 20 | 69.80 | Leather, % of EBW | 3.66 × 10−7 | 9 | 69.69–69.93 | ** |
| 22 | 24.59 | Rumen, % of EBW | 2.77 × 10−6 | 10 | 24.56–24.60 | ** |
| 26 | 43.11 | Intestines, % of EBW | 1.09 × 10−6 | 2 | 43.10–43.11 | ** |
| 28 | 25.43 | REI maize phase | 6.43 × 10−8 | 16 | 25.41–25.55 | *** |
| 28 | 28.24 | Intestines, % of EBW | 7.77 × 10−7 | 13 | 28.24–28.37 | ** |
1*** corresponds to the genome-wide significance threshold and ** to the chromosome-wide significance threshold.
All but one of the QTLs significant at a genome-wide level were located on the Bos Taurus chromosome (BTA) 2. These QTLs, associated with carcass yield, fifth quarter proportion, fifth quarter fat, percentage of muscle, percentage of fat, and liver proportion, were all located at 6.2 Mb. The QTLs were also significant at a chromosome-wide level for intestine proportion, digestive tract proportion, and leather proportion. The level of association was extremely high for several traits: fifth quarter proportion (−log P = 28), carcass yield (−log P = 23), percentage of muscle (−log P = 25), and percentage of fat (−log P = 15). The minor allele frequency of the SNPmax was 0.13.
The last QTL to reach the threshold of genome-wide significance was associated with the REI during the maize phase and located on BTA 28 at 25 Mb. The frequency of the minor allele was 0.28 and the SNP effect corresponded to a 0.38 phenotypic SD.
DISCUSSION
REI Modeling
This study used a very classic REI model with fixed effects similar to those in the literature. However, the R2 values of our models (0.15 and 0.49) were low, particularly for the hay phase model. In their review, Kenny et al. (2018) reported that most studies on growing beef cattle under an energy-dense diet (e.g., Kelly et al., 2010; Fitzsimons et al., 2014) indicated an R2 of around 0.7. When the diet mainly contained forage, the R2 reported in the literature is slightly lower at around 0.6 (e.g., Shaffer et al., 2011; Lawrence et al., 2012). This difference between diets has been hypothesized as being due to differences in the rumen fill value that might influence the DM intake (DMI) (Kelly et al., 2010; Kenny et al., 2018). In our specific case, we cumulated two parameters that could explain the low R2 value: first, the animals were fed with forage-based diets and, second, the study was performed on adult, full-grown animals so that changes to body weight only reflected changes in body reserves. The small variation in body weight observed during our study, particularly during the hay phase, was logically a factor limiting the importance of the ADG in the R2 of the model when compared to growing animals. Indeed, the ADG effect was not significant in the hay phase model. Lawrence et al. (2013), who studied pregnant nongrowing beef cows fed with grass silage, also found a low R2 value (R2 = 0.24).
Genetic Parameters for Feed Efficiency
The heritability estimates for REI were quite low (0.12) but similar between the two phases. However, the SE of 0.08 was quite large and suggests that the true heritability could be between 0 (included) and 0.28. A higher number of animals would have helped to improve the accuracy of our estimation.
This heritability value of 0.12 was within the lower range of heritability for RFI reported in the literature, as reviewed by Berry and Crowley (2013). Indeed, they saw heritability values ranging from 0.00 to 0.62. However, heritability varies between studies depending on the type of animal involved. Although the range reported by Berry and Crowley (2013) for growing animals is between 0.14 and 0.62, this falls to between 0.00 and 0.38 in adult cows, which is in line with our estimates. This discrepancy between the heritability values of growing and adult animals may indicate that the biological processes hidden behind the feed efficiency trait are at least in part due to differences in their developmental stage. Indeed, the proportion of maintenance on the total energy requirements differs markedly between growing animals and their adult counterparts. In other words, one might say that an efficient use of feed for growth is more heritable than an efficient use of feed for maintenance. Heritability estimates relative to body weight and energy intake were similar in the two phases and in line with data in the literature (Berry and Crowley, 2013). However, the ADG estimates were lower than those reported, probably, because the latter were all estimated in growing animals.
As expected due to the regression properties, phenotypic correlations of REI with energy intake were positive and high for both phases. This result was consistent with the literature reviewed by Berry and Crowley (2013), which had reported an average phenotypic correlation of 0.66 between RFI and feed intake. Other studies also reported similar phenotypic correlations (e.g., 0.72 for Coyne et al., 2018 and 0.65 for Polizel et al., 2018). The genetic correlations of 0.88 and 0.68 found between REI and energy intake for the two different phases were also consistent with equivalent correlations in the literature: 0.72, 0.61, and 0.70 according to Berry and Crowley (2013), Coyne et al. (2018), and Polizel et al. (2018), respectively. Phenotypic correlations of REI with weight and ADG were 0 by construction, and none of the genetic correlations differed from 0 in this study. This result is similar to most of the findings reported in the literature (Berry and Crowley, 2013), although some studies did find correlations (e.g., Nkrumah et al., 2007a; Polizel et al., 2018).
REI Under Two Different Diets
The genetic correlation between the REI values during the two phases was high (0.83), indicating that, although they were two distinct traits, they are, nevertheless, markedly similar from a genetic point of view. Some studies had previously explored the effect of different diets on feed efficiency but mostly from the phenotypic point of view. Durunna et al. (2011a) and Cassady et al. (2016) found moderate phenotypic correlations between the RFI of young cattle determined under both growing and finishing diets (0.33 and 0.40, respectively). Manafiazar et al. (2015) reported a correlation of 0.30 for RFI between drylot conditions and pasture. By contrast, other studies did not find any difference in RFI on pasture among animals ranked according to their RFI evaluated indoors (Lawrence et al., 2012; Oliveira et al., 2016). Finally, Coyle et al. (2016, 2017) estimated correlations of 0.18 and 0.19 between RFI on distributed fresh grass diet and RFI on high concentrate diet and correlations of 0.30 and 0.40 between RFI on grass silage and RFI on distributed fresh grass in two different breeds of cattle. All these phenotypic correlations were within the same range (between 0.2 and 0.4), which also covers the phenotypic correlation estimated during our study (0.36). The study by Durunna et al. (2011b) was one of the rare works that published a genetic correlation in this context, finding an estimated genetic correlation of 0.50 between RFI on growing and finishing diets. Although the genetic correlation we estimated was higher, these published correlations were both high as well, suggesting the presence of genetic × environment (G × E) interactions. Although estimates of genetic correlations are still rare in that case, a consensus seems to have been reached in the recent literature concerning the existence of G × E interactions with respect to feed efficiency (Berry and Crowley, 2013; Cantalapiedra-Hijar et al., 2018; Kenny et al., 2018). From a biological point of view, this interaction may be due to both differences in feeding behavior and digestive efficiency. Difference in the rumen microbiota between efficient and nonefficient animals, in interaction with the diet, have also been demonstrated (Zhou et al., 2010; Carberry et al., 2012; Hernandez-Sanabria et al., 2012).
Slaughter Traits and Their Links with Feed Efficiency
Slaughter traits have been found as being moderately heritable. Ríos Utrera and Van Vleck (2004) reviewed 72 papers from the literature and reported the heritability of several carcass traits. Although our estimates were always within the range of those reported, they differed slightly from the overall mean. The review reported an average heritability of 0.40 for hot carcass weight, which was lower than the 0.68 we estimated here for EBW (although this is not exactly the same trait). Similarly, our estimate was higher than the average for carcass yield (0.53 vs. 0.32), while the heritability of muscle percentage and fat percentage were lower (0.33 vs. 0.52 and 0.27 vs. 0.51, respectively). The values for fifth quarter fat were close (0.33 vs. 0.40). Previous studies on French Charolais cattle had reported values close to our estimates: 0.43, 0.54, and 0.68 for carcass yield (Renand, 1985; Fouilloux et al., 1999; Renand and Krauss, 2002) and 0.41 and 0.36 for muscle and fat percentage, respectively (Renand and Krauss, 2002). The latter authors also estimated the heritability of three visceral organ traits. They found a higher heritability than our estimates for the digestive tract (0.45 vs. 0.20) and lung (0.39 vs. 0.05) and a lower heritability for the heart (0.23 vs. 0.50). Although some authors have suggested that visceral organ masses may be heritable (Jenkins et al., 1986; Hotovy et al., 1991), we were not able to find any other estimates of visceral organ weight heritability in cattle.
We found low phenotypic correlations between REI and some carcass traits, suggesting that efficient animals have generally a slightly higher muscle percentage and lower fat percentage in their carcasses compared to high REI animals and that a larger proportion of EBW is taken in high REI animals by digestive organs and the liver when compared to efficient animals. Some studies in the literature did not find any phenotypic correlations between RFI and most carcass composition or organ weight traits (e.g., Mader et al., 2009; Cruz et al., 2010; Fitzsimons et al., 2014; Fidelis et al., 2017). However, when correlations were found, they were in line with those observed during our study. A link between a low RFI and less internal fat was reported by Richardson et al. (2001) and Mader et al. (2009). Fitzsimons et al. (2014) also found differences in the digestive tract between low and high RFI groups of animals, with efficient animals having the lightest reticulo-rumen. Basarab et al. (2003) and Bonilha et al. (2013) found a similar trend regarding both the gastrointestinal tract and liver.
We found similar correlations at the genetic level, although only a few were significant because of large SEs. Once again, the results were in line with the genetic correlations found in the literature. For instance, Nkrumah et al. (2007a) found a negative genetic relation between RFI and both carcass lean meat area and lean meat yield, while the correlation with carcass grade fat was positive. Similar results were also observed by Robinson and Oddy (2004), Crowley et al. (2011), and Coyne et al. (2018). In their meta-analysis, Berry and Crowley (2013) reported genetic correlations of −0.18 between lean and RFI and of 0.20 between fat and RFI. To our knowledge, there are few data in the literature on genetic correlations between visceral organ weight and RFI. Nevertheless, a study by Renand and Krauss (2002) reported a positive genetic correlation of 0.42 between RFI and the empty digestive tract in young Charolais bulls.
The link between feed efficiency and carcass composition, with more muscle and less fat deposition among efficient animals, accords with the fact that the synthesis of protein is energetically more efficient than that of fat (Archer et al., 1999; Cantalapiedra-Hijar et al., 2018). However, the same studies also mentioned that, in contrast, the maintenance of fat required less energy than that of protein, which makes this finding difficult to interpret from a biological point of view. As for the relationship between feed efficiency and gastrointestinal and liver weight or proportion, the interpretation is easier and supported biologically. First, it has been reported that fluctuations in the liver and gastrointestinal tract appear to be directly proportional to dietary intake (Johnson et al., 1990; Ortigues and Doreau, 1995; Archer et al., 1999), with feed-restricted animals displaying a reduction in their organ weights or proportions. Second, the digestive tract and liver are among the most metabolically active tissues (Smith and Baldwin, 1974; Ferrell and Jenkins, 1985). Indeed, it has been reported that the liver accounts for 25% of energy expenditure in steers, while the gut represents 23% (Lobley, 2003), and their protein turnover has been found to be higher than that of skeletal muscles (Early et al., 1990). It has also been suggested that individual differences in protein turnover rate may be a component of feed efficiency (Archer et al., 1999; Fitzsimons et al., 2014; Cantalapiedra-Hijar et al., 2018).
Association Analysis
The broad effect of the myostatin gene.
As mentioned above, almost all the QTLs significant at the genome-wide level, and some at the chromosome-wide level, were located on BTA 2. These QTLs correspond to the effect of the myostatin gene which was identified as the GDF8 gene and is localized in the start of BTA 2 (Charlier et al., 1995; Grobet et al., 1997; Grobet et al., 1998). This gene regulates muscle fiber deposition and its mutations are now acknowledged as causing the double-muscling phenotype, with considerable consequences in different species (Bellinge et al., 2005). Here, we found a strong effect of this gene on carcass traits such as carcass yield, fat percentage, and protein percentage, in line with the abundant literature on this subject (e.g., Allais et al., 2010; Martínez et al., 2010; Morris et al., 2010; Sorbolini et al., 2017). In addition, we found that this gene also affected some organ proportions: intestine, digestive tract, and liver, as well as leather and internal fat. This effect on internal organ proportions had already been observed by Morris et al. (2010) with respect to the liver, digestive tract, and internal fat. They also reported an effect on lung proportion that we did not observe. Furthermore, an association between the double-muscling phenotype and heart size has been reported in the past (Monin et al., 1974), but neither Morris et al. nor our team identified any QTL supporting this observation, despite the fact that a similar effect has been found in mice (Bünger et al., 2004).
Feed efficiency—is a gene family implicated?
Among all the efficiency traits on which GWAS was performed, only the REI estimated during the maize phase produced significant results. Numerous studies have reported QTLs for RFI in recent years (e.g., Nkrumah et al., 2007b; Sherman et al., 2010; Rolf et al., 2012; Saatchi et al., 2014; Seabury et al., 2017). However, the results of these GWAS were generally inconsistent, with different QTLs being found from one study to another. To date, there have been no fewer than 494 reports for RFI and 134 reports for feed conversion traits in the cattle QTL database (https://www.animalgenome.org/cgi-bin/QTLdb/index). The results of our study were not an exception to this rule, with the QTLs found on BTA 13 being in a region that has never previously been reported. The nearest QTL in the literature, detected for maintenance efficiency, efficiency of gain, and partial efficiency of growth by de Oliveira et al. (2014), is indeed at a distance of 10 Mb. The maximum of our QTL peak was located in an intergenic region, the nearest genes being MACROD2, FLRT3, and KIF16B. The second significant QTL that we detected for REI on BTA 19 is located less than 1 Mb away from a QTL that was previously reported by Nkrumah et al. (2007b) for RFI and Rolf et al. (2012) for feed intake. The maximum of our QTL peak is an intronic SNP of the TANC2 gene. Although the biological functions of the protein coded by the TANC2 gene remain unclear, it has been associated in the literature with neurons and embryonic development (Han et al., 2010). The last QTL that we identified for REI was the most significant and found on BTA 28. Our QTL co-localized with two QTLs for DMI that had been identified by Hardie et al. (2017) and Tetens et al. (2014). It is worth noting that the QTL reported by Hardie et al. (2017) was found in their multiparous but not primiparous cow populations. Tetens et al. (2014) reported that their QTL region contained a cluster of olfactory receptor genes that might be related to intake. However, the maximum of our QTL peak appeared to be an intronic SNP of the KIF1BP gene, also called KBP. This kinesin binding protein, mainly localized in mitochondria, interacts with the cytoskeleton, and plays a role in nervous system development (Wozniak et al., 2005; Alves et al., 2010). The KBP protein is known to interact with different KIF1 proteins (including KIF1A) and could be a factor governing their regulation (Wozniak et al., 2005). This result is particularly interesting because the TANC2 protein associated with the QTL found on BTA 19 is also known to interact with the KIF1A protein (Stucchi et al., 2018). It is also noteworthy that among the genes close to our QTL on BTA 13 is KIF16B, which also belongs to the kinesin superfamily and shares a high degree of homology with KIF1A regarding its active domains (Lawrence et al., 2004). However, although there was some concordance among the genes found under our QTL peaks and their associated pathways, no obvious links could be made between this metabolic pathway and the biology of feed efficiency.
Other slaughter QTL—a strong candidate gene to determine liver size.
Apart from the QTL on BTA 2 associated with GDF8 described above, all the other significant QTLs detected during our study for slaughter traits concerned organ proportions. To our knowledge, only two other publications have explored QTLs for these traits (Morris et al., 2010; An et al., 2018). However, none of our QTLs was common to those previously reported in the literature. The small number of animals (several hundred in our study and that by Morris et al. and around a thousand in that by An et al.) and the fact that these studies were performed on different breeds did not facilitate the statistic power of the analyses and any potential convergence of the results regarding these moderately heritable traits. All the genes localized in close proximity to our detected QTL were examined. Two of them were particularly interesting. The first is the SLC16A1 gene, which is localized 3 kb from our QTL peak for Abomasum on BTA 3 and is involved in energy metabolism (Halestrap, 2013), while the second is the WASL gene, found in close proximity to our QTL for liver proportion on BTA 4. This gene has been reported to be differentially expressed in cancerous human liver cells versus noncancerous cells (Costantini et al., 2013). Moreover, this gene has also been found to be overexpressed in the liver of cattle during the refeeding phase after feed restriction (Connor et al., 2010), and these authors also reported a link between feed restriction and a reduction in liver size. The WASL gene is, therefore, a strong candidate to exert a potential genetic effect on liver size, as well as having a link with feed efficiency, as previously shown.
No QTL was found to be common to REI and slaughter traits despite some very high genetic correlations. Similarly, there was no common QTL between the REI of the two diet phases. Moreover, there was no significant QTL for REI in the hay phase. It is true that the heritability of REI is low but it is also likely that the small number of animals tested during the study restricted our detection power.
As has already been noted, our work involved an unusual type of animal when studying feed efficiency in beef cattle: adult cows receiving forage diets. Most studies in the literature analyzed RFI in growing (finishing) animals supplied with energy-dense diets. This affected the conduct of our study. For instance, we chose only to use residual intake as a feed efficiency trait and to discard other possible traits, such as the feed conversion ratio or residual gain, because of the small variations in body weight observed in our full-grown animals. Similarly, it may not be possible to directly compare our results with those in the traditional literature because of these differences in both life stage and diet. Although we have mentioned the G × E effect observed with the diet, some studies have also investigated the relationship with life stage (e.g., Nieuwhof et al., 1992; Gomes et al., 2012) and also suggested a G × E effect. This raises the question of the optimum selection goal in beef cattle in regards with feed efficiency: should this be young animals fed a concentrate-based diet, a reproductive herd raised on a forage-based or grass-based diet, or should we try to identify common foundations for efficiency between these different types of feed and animal? Further investigations are, therefore, necessary in this respect.
ACKNOWLEDGMENTS
The authors would like to thank the technical staff at the INRA experimental slaughterhouse, 63122 Saint-Genès-Champanelle, France.
Conflict of interest statement. The authors have no conflicts of interest to declare.
REFERENCES
- Allais S., Levéziel H., Payet-Duprat N., Hocquette J. F., Lepetit J., Rousset S., Denoyelle C., Bernard-Capel C., Journaux L., Bonnot A., et al. . 2010. The two mutations, Q204X and nt821, of the myostatin gene affect carcass and meat quality in young heterozygous bulls of french beef breeds. J. Anim. Sci. 88:446–454. doi: 10.2527/jas.2009-2385 [DOI] [PubMed] [Google Scholar]
- Alves M. M., Burzynski G., Delalande J. M., Osinga J., van der Goot A., Dolga A. M., de Graaff E., Brooks A. S., Metzger M., Eisel U. L., et al. . 2010. KBP interacts with SCG10, linking goldberg-shprintzen syndrome to microtubule dynamics and neuronal differentiation. Hum. Mol. Genet. 19:3642–3651. doi: 10.1093/hmg/ddq280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B., Xia J., Chang T., Wang X., Miao J., Xu L., Zhang L., Gao X., Chen Y., Li J., et al. . 2018. Genome-wide association study identifies loci and candidate genes for internal organ weights in simmental beef cattle. Physiol. Genomics. 50:523–531. doi: 10.1152/physiolgenomics.00022.2018 [DOI] [PubMed] [Google Scholar]
- Archer J. A., Richardson E. C., Herd R. M., and Arthur P. F.. 1999. Potential for selection to improve efficiency of feed use in beef cattle: a review. Aust. J. Agric. Res. 50:147–162. doi: 10.1071/a98075 [DOI] [Google Scholar]
- Basarab J. A., Price M. A., Aalhus J. L., Okine E. K., Snelling W. M., and Lyle K. L.. 2003. Residual feed intake and body composition in young growing cattle. Can. J. Anim. Sci. 83:189–204. doi: 10.4141/A02-065 [DOI] [Google Scholar]
- Bellinge R. H., Liberles D. A., Iaschi S. P., O’brien P. A., and Tay G. K.. 2005. Myostatin and its implications on animal breeding: a review. Anim. Genet. 36:1–6. doi: 10.1111/j.1365-2052.2004.01229.x [DOI] [PubMed] [Google Scholar]
- Berry D. P., and Crowley J. J.. 2013. Cell biology symposium: genetics of feed efficiency in dairy and beef cattle. J. Anim. Sci. 91:1594–1613. doi: 10.2527/jas.2012-5862 [DOI] [PubMed] [Google Scholar]
- Boichard D., Guillaume F., Baur A., Croiseau P., Rossignol M. N., Boscher M. Y., Druet T., Genestout L., Colleau J. J., Journaux L., et al. . 2012. Genomic selection in French dairy cattle. Anim. Prod. Sci. 52:115–120. doi: 10.1071/AN11119 [DOI] [Google Scholar]
- Bonilha E. F., Branco R. H., Bonilha S. F., Araujo F. L., Magnani E., and Mercadante M. E.. 2013. Body chemical composition of nellore bulls with different residual feed intakes. J. Anim. Sci. 91:3457–3464. doi: 10.2527/jas.2012-5437 [DOI] [PubMed] [Google Scholar]
- Bünger L., Ott G., Varga L., Schlote W., Rehfeldt C., Renne U., Williams J. L., and Hill W. G.. 2004. Marker-assisted introgression of the compact mutant myostatin allele MstnCmpt-dl1Abc into a mouse line with extreme growth effects on body composition and muscularity. Genet. Res. 84:161–173. doi:10.1017/S0016672304007165 [DOI] [PubMed] [Google Scholar]
- Campo J. L., and Rodríguez M.. 1990. Relative efficiency of selection methods to improve a ratio of two traits in tribolium. Theor. Appl. Genet. 80:343–348. doi: 10.1007/BF00210070 [DOI] [PubMed] [Google Scholar]
- Cantalapiedra-Hijar G., Abo-Ismail M., Carstens G. E., Guan L. L., Hegarty R., Kenny D. A., McGee M., Plastow G., Relling A., and Ortigues-Marty I.. 2018. Review: biological determinants of between-animal variation in feed efficiency of growing beef cattle. Animal. 12(s2):s321–s335. doi: 10.1017/S1751731118001489 [DOI] [PubMed] [Google Scholar]
- Carberry C. A., Kenny D. A., Han S., McCabe M. S., and Waters S. M.. 2012. Effect of phenotypic residual feed intake and dietary forage content on the rumen microbial community of beef cattle. Appl. Environ. Microbiol. 78:4949–4958. doi: 10.1128/AEM.07759-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassady C. J., Felix T. L., Beever J. E., and Shike D. W.. 2016. Effects of timing and duration of test period and diet type on intake and feed efficiency of charolais-sired cattle. J. Anim. Sci. 94:4748–4758. doi: 10.2527/jas.2016-0633 [DOI] [PubMed] [Google Scholar]
- Charlier C., Coppieters W., Farnir F., Grobet L., Leroy P. L., Michaux C., Mni M., Schwers A., Vanmanshoven P., and Hanset R.. 1995. The mh gene causing double-muscling in cattle maps to bovine chromosome 2. Mamm. Genome. 6:788–792. doi:10.1007/BF00539005 [DOI] [PubMed] [Google Scholar]
- Connor E. E., Kahl S., Elsasser T. H., Parker J. S., Li R. W., Van Tassell C. P., Baldwin R. L. 6th, and Barao S. M.. 2010. Enhanced mitochondrial complex gene function and reduced liver size may mediate improved feed efficiency of beef cattle during compensatory growth. Funct. Integr. Genomics. 10:39–51. doi: 10.1007/s10142-009-0138-7 [DOI] [PubMed] [Google Scholar]
- Costantini S., Di Bernardo G., Cammarota M., Castello G., and Colonna G.. 2013. Gene expression signature of human HepG2 cell line. Gene. 518:335–345. doi: 10.1016/j.gene.2012.12.106 [DOI] [PubMed] [Google Scholar]
- Coyle S., Fitzsimons C., Kenny D. A., and Kelly A.. 2017. Feed efficiency correlations in beef cattle offered zero-grazed grass and a high-concentrate diet. Adv. Anim. Biosci. 8:121. [Google Scholar]
- Coyle S., Fitzsimons C., Kenny D. A., Kelly A. K., and McGee M.. 2016. Repeatability of feed efficiency in beef cattle offered grass silage and zero-grazed grass. J. Anim. Sci. 94:719–719. doi: 10.2527/jam2016-1481 [DOI] [Google Scholar]
- Coyne J. M., Judge M. M., Conroy S., and Berry D. P.. 2018. Variance component estimation of efficiency, carcass and meat quality traits in beef cattle. In: Proceedings of the World Congress on Genetics Applied to Livestock Production Digital Archive; WCGALP 2018, February 11–16, Auckland, New Zealand Volume: Electronic Poster Session—Biology—Feed Intake and Efficiency 1. p. 912. [Google Scholar]
- Crowley J. J., Evans R. D., Mc Hugh N., Pabiou T., Kenny D. A., McGee M., Crews D. H., and Berry D. P.. 2011. Genetic associations between feed efficiency measured in a performance test station and performance of growing cattle in commercial beef herds. J. Anim. Sci. 89:3382–3393. doi: 10.2527/jas.2011-3836 [DOI] [PubMed] [Google Scholar]
- Cruz G. D., Rodríguez-Sánchez J. A., Oltjen J. W., and Sainz R. D.. 2010. Performance, residual feed intake, digestibility, carcass traits, and profitability of angus-hereford steers housed in individual or group pens. J. Anim. Sci. 88:324–329. doi: 10.2527/jas.2009-1932 [DOI] [PubMed] [Google Scholar]
- Durunna O. N., Mujibi F. D. N., Goonewardene L., Okine E. K., Basarab J. A., Wang Z., and Moore S. S.. 2011a. Feed efficiency differences and reranking in beef steers fed grower and finisher diets. J. Anim. Sci. 89:158–167. doi: 10.2527/jas.2009-2514 [DOI] [PubMed] [Google Scholar]
- Durunna O. N., Plastow G., Mujibi F. D. N., Grant J., Mah J., Basarab J. A., Okine E. K., Moore S. S., and Wang Z.. 2011b. Genetic parameters and genotype × environment interaction for feed efficiency traits in steers fed grower and finisher diets. J. Anim. Sci. 89:3394–3400. doi: 10.2527/jas.2010-3516 [DOI] [PubMed] [Google Scholar]
- Early R. J., McBride B. W., and Ball R. O.. 1990. Growth and metabolism in somatotropin-treated steers: III. Protein synthesis and tissue energy expenditures. J. Anim. Sci. 68:4153–4166. doi: 10.2527/1990.68124153x [DOI] [PubMed] [Google Scholar]
- Ferrell C. L., and Jenkins T. G.. 1985. Cow type and the nutritional environment: nutritional aspects. J. Anim. Sci. 61:725–741. doi: 10.2527/jas1985.613725x [DOI] [PubMed] [Google Scholar]
- Fidelis H. A., Bonilha S. F. M., Tedeschi L. O., Branco R. H., Cyrillo J. N. S. G., and Mercadante M. E. Z.. 2017. Residual feed intake, carcass traits and meat quality in nellore cattle. Meat Sci. 128:34–39. doi: 10.1016/j.meatsci.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Fitzsimons C., Kenny D. A., and McGee M.. 2014. Visceral organ weights, digestion and carcass characteristics of beef bulls differing in residual feed intake offered a high concentrate diet. Animal. 8:949–959. doi: 10.1017/S1751731114000652 [DOI] [PubMed] [Google Scholar]
- Fouilloux M.-N., Renand G., Gaillard J., and Ménissier F.. 1999. Genetic parameters of beef traits of limousin and charolais progeny-tested AI sires. Genet. Sel. Evol. 31:465. doi: 10.1186/1297-9686-31-5-465 [DOI] [Google Scholar]
- Gomes R. C., Sainz R. D., Silva S. L., César M. C., Bonin M. N., and Leme P. R.. 2012. Feedlot performance, feed efficiency reranking, carcass traits, body composition, energy requirements, meat quality and calpain system activity in Nellore steers with low and high residual feed intake. Livest. Sci. 150:265–273. doi: 10.1016/j.livsci.2012.09.012 [DOI] [Google Scholar]
- Grobet L., Martin L. J., Poncelet D., Pirottin D., Brouwers B., Riquet J., Schoeberlein A., Dunner S., Ménissier F., Massabanda J., et al. . 1997. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 17:71–74. doi: 10.1038/ng0997-71 [DOI] [PubMed] [Google Scholar]
- Grobet L., Poncelet D., Royo L. J., Brouwers B., Pirottin D., Michaux C., Ménissier F., Zanotti M., Dunner S., and Georges M.. 1998. Molecular definition of an allelic series of mutations disrupting the myostatin function and causing double-muscling in cattle. Mamm. Genome. 9:210–213. doi:10.1007/s003359900727 [DOI] [PubMed] [Google Scholar]
- Gunsett F. C. 1984. Linear index selection to improve traits defined as ratios. J. Anim. Sci. 59:1185–1193. doi: 10.2527/jas1984.5951185x [DOI] [Google Scholar]
- Halestrap A. P. 2013. The SLC16 gene family— structure, role and regulation in health and disease. Mol. Aspects Med. 34:337–349. doi: 10.1016/j.mam.2012.05.003 [DOI] [PubMed] [Google Scholar]
- Han S., Nam J., Li Y., Kim S., Cho S. H., Cho Y. S., Choi S. Y., Choi J., Han K., Kim Y., et al. . 2010. Regulation of dendritic spines, spatial memory, and embryonic development by the TANC family of PSD-95-interacting proteins. J. Neurosci. 30:15102–15112. doi: 10.1523/JNEUROSCI.3128-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie L. C., VandeHaar M. J., Tempelman R. J., Weigel K. A., Armentano L. E., Wiggans G. R., Veerkamp R. F., de Haas Y., Coffey M. P., Connor E. E., et al. . 2017. The genetic and biological basis of feed efficiency in mid-lactation holstein dairy cows. J. Dairy Sci. 100:9061–9075. doi: 10.3168/jds.2017-12604 [DOI] [PubMed] [Google Scholar]
- Herd R. M., Oddy V. H., and Richardson E. C.. 2004. Biological basis for variation in residual feed intake in beef cattle. 1. Review of potential mechanisms. Aust. J. Exp. Agric. 44:423–430. doi: 10.1071/ea02220 [DOI] [Google Scholar]
- Hernandez-Sanabria E., Goonewardene L. A., Wang Z., Durunna O. N., Moore S. S., and Guan L. L.. 2012. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 78:1203–1214. doi: 10.1128/AEM.05114-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotovy S. K., Johnson K. A., Johnson D. E., Carstens G. E., Bourdon R. M., and Seidel G. E. Jr. 1991. Variation among twin beef cattle in maintenance energy requirements. J. Anim. Sci. 69:940–946. doi: 10.2527/1991.693940x [DOI] [PubMed] [Google Scholar]
- Hozé C., Fouilloux M. N., Venot E., Guillaume F., Dassonneville R., Fritz S., Ducrocq V., Phocas F., Boichard D., and Croiseau P.. 2013. High-density marker imputation accuracy in sixteen french cattle breeds. Genet. Sel. Evol. 45:33. doi: 10.1186/1297-9686-45-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institut National de la Recherche Agronomique 2018. Alimentation des ruminants. Apports nutritionnels—Besoin et réponses des animaux—Rationnement—Tables des valeurs des aliments. Versailles (France): Editions Quae. [Google Scholar]
- Jenkins T. G., Ferrell C. L., and Cundiff L. V.. 1986. Relationship of components of the body among mature cows as related to size, lactation potential and possible effects on productivity. Anim. Sci. 43:245–254. doi: 10.1017/S0003356100002427 [DOI] [Google Scholar]
- Johnson D. E., Johnson K. A., and Baldwin R. L.. 1990. Changes in liver and gastrointestinal tract energy demands in response to physiological workload in ruminants. J. Nutr. 120:649–655. doi: 10.1093/jn/120.6.649 [DOI] [PubMed] [Google Scholar]
- Kelly A. K., McGee M., Crews D. H., Fahey A. G., Wylie A. R., and Kenny D. A.. 2010. Effect of divergence in residual feed intake on feeding behavior, blood metabolic variables, and body composition traits in growing beef heifers. J. Anim. Sci. 88:109–123. doi: 10.2527/jas.2009-2196 [DOI] [PubMed] [Google Scholar]
- Kenny D. A., Fitzsimons C., Waters S. M., and McGee M.. 2018. Invited review: improving feed efficiency of beef cattle - the current state of the art and future challenges. Animal. 12:1815–1826. doi: 10.1017/S1751731118000976 [DOI] [PubMed] [Google Scholar]
- Khiaosa-ard R., and Zebeli Q.. 2014. Cattle’s variation in rumen ecology and metabolism and its contributions to feed efficiency. Livest. Sci. 162:66–75. doi: 10.1016/j.livsci.2014.01.005 [DOI] [Google Scholar]
- Klosterman E. W. 1972. Beef cattle size for maximum efficiency. J. Anim. Sci. 34:875–880. doi: 10.2527/jas1972.345875x [DOI] [Google Scholar]
- Koch R. M., Swiger L. A., Chambers D., and Gregory K. E.. 1963. Efficiency of feed use in beef cattle. J. Anim. Sci. 22:486–494. doi: 10.2527/jas1963.222486x [DOI] [Google Scholar]
- Lawrence C. J., Dawe R. K., Christie K. R., Cleveland D. W., Dawson S. C., Endow S. A., Goldstein L. S. B., Goodson H. V., Hirokawa N., Howard J., et al. . 2004. A standardized kinesin nomenclature. J. Cell Biol. 167:19–22. doi: 10.1083/jcb.200408113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P., Kenny D. A., Earley B., and McGee M.. 2012. Grazed grass herbage intake and performance of beef heifers with predetermined phenotypic residual feed intake classification. Animal. 6:1648–1661. doi: 10.1017/S1751731112000559 [DOI] [PubMed] [Google Scholar]
- Lawrence P., Kenny D. A., Earley B., and McGee M.. 2013. Intake of conserved and grazed grass and performance traits in beef suckler cows differing in phenotypic residual feed intake. Livest. Sci. 152:154–166. doi: 10.1016/j.livsci.2012.12.024 [DOI] [Google Scholar]
- Lobley G. E. 2003. Protein turnover—what does it mean for animal production? Can. J. Anim. Sci. 83:327–340. doi: 10.4141/A03-019 [DOI] [Google Scholar]
- Lu D., Miller S., Sargolzaei M., Kelly M., Vander Voort G., Caldwell T., Wang Z., Plastow G., and Moore S.. 2013. Genome-wide association analyses for growth and feed efficiency traits in beef cattle. J. Anim. Sci. 91:3612–3633. doi: 10.2527/jas.2012-5716 [DOI] [PubMed] [Google Scholar]
- Mader C. J., Montanholi Y. R., Wang Y. J., Miller S. P., Mandell I. B., McBride B. W., and Swanson K. C.. 2009. Relationships among measures of growth performance and efficiency with carcass traits, visceral organ mass, and pancreatic digestive enzymes in feedlot cattle. J. Anim. Sci. 87:1548–1557. doi: 10.2527/jas.2008-0914 [DOI] [PubMed] [Google Scholar]
- Manafiazar G., Basarab J. A., Baron V. S., McKeown L., Doce R. R., Swift M., Undi M., Wittenberg K., and Ominski K.. 2015. Effect of post-weaning residual feed intake classification on grazed grass intake and performance in pregnant beef heifers. Can. J. Anim. Sci. 95:369–381. doi: 10.4141/cjas-2014-184 [DOI] [Google Scholar]
- Martínez A., Aldai N., Celaya R., and Osoro K.. 2010. Effect of breed body size and the muscular hypertrophy gene in the production and carcass traits of concentrate-finished yearling bulls. J. Anim. Sci. 88:1229–1239. doi: 10.2527/jas.2009-2025 [DOI] [PubMed] [Google Scholar]
- Meyer K. 2007. WOMBAT: a tool for mixed model analyses in quantitative genetics by restricted maximum likelihood (REML). J. Zhejiang Univ. Sci. B. 8:815–821. doi: 10.1631/jzus.2007.B0815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monin G., Boccard R., Vernin P., Talmant A., and Nicolas N.. 1974. Caractéristiques physiologiques respiratoires des bovins culards. Ann. Genet. Sel. Anim. 6:187–193. doi: 10.1186/1297-9686-6-2-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaño-Bermudez M., Nielsen M. K., and Deutscher G. H.. 1990. Energy requirements for maintenance of crossbred beef cattle with different genetic potential for milk. J. Anim. Sci. 68:2279–2288. doi: 10.2527/1990.6882279x [DOI] [PubMed] [Google Scholar]
- Morris C. A., Bottema C. D., Cullen N. G., Hickey S. M., Esmailizadeh A. K., Siebert B. D., and Pitchford W. S.. 2010. Quantitative trait loci for organ weights and adipose fat composition in jersey and limousin back-cross cattle finished on pasture or feedlot. Anim. Genet. 41:589–596. doi: 10.1111/j.1365-2052.2010.02058.x [DOI] [PubMed] [Google Scholar]
- Nieuwhof G. J., van Arendonk J. A. M., Vos H., and Korver S.. 1992. Genetic relationships between feed intake, efficiency and production traits in growing bulls, growing heifers and lactating heifers. Livest. Prod. Sci. 32:189–202. doi: 10.1016/S0301-6226(12)80001-7 [DOI] [Google Scholar]
- Nkrumah J. D., Basarab J. A., Wang Z., Li C., Price M. A., Okine E. K., Crews D. H., and Moore S. S.. 2007a. Genetic and phenotypic relationships of feed intake and measures of efficiency with growth and carcass merit of beef cattle. J. Anim. Sci. 85:2711–2720. doi: 10.2527/jas.2006-767 [DOI] [PubMed] [Google Scholar]
- Nkrumah J. D., Sherman E. L., Li C., Marques E., Crews D. H. Jr, Bartusiak R., Murdoch B., Wang Z., Basarab J. A., and Moore S. S.. 2007b. Primary genome scan to identify putative quantitative trait loci for feedlot growth rate, feed intake, and feed efficiency of beef cattle. J. Anim. Sci. 85:3170–3181. doi: 10.2527/jas.2007-0234 [DOI] [PubMed] [Google Scholar]
- de Oliveira P. S., Cesar A. S., do Nascimento M. L., Chaves A. S., Tizioto P. C., Tullio R. R., Lanna D. P., Rosa A. N., Sonstegard T. S., Mourao G. B., et al. . 2014. Identification of genomic regions associated with feed efficiency in nelore cattle. BMC Genet. 15:100. doi: 10.1186/s12863-014-0100-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira L. F., Ruggieri A. C., Branco R. H., Cota O. L., Canesin R. C., Costa H. J. U., and Mercadante M. E. Z.. 2016. Feed efficiency and enteric methane production of Nellore cattle in the feedlot and on pasture. Anim. Prod. Sci. 58:886–893. doi: 10.1071/AN16303 [DOI] [Google Scholar]
- Ortigues I., and Doreau M.. 1995. Responses of the splanchnic tissues of ruminants to changes in intake : absorption of digestion end products, tissue mass, metabolic activity and implications to whole animal energy metabolism. Ann. Zootech. 44:321–346. [Google Scholar]
- Polizel G. H. G., Grigoletto L., Carvalho M. E., Rossi Junior P., Ferraz J. B. S., and de A. Santana M. H.. 2018. Genetic correlations and heritability estimates for dry matter intake, weight gain and feed efficiency of Nellore cattle in feedlot. Livest. Sci. 214:209–210. doi: 10.1016/j.livsci.2018.06.013 [DOI] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J., et al. . 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renand G. 1985. Genetic parameters of french beef breeds used in crossbreeding for young bull production. II.—slaughter performance. Genet. Sel. Evol. 17:265–282. doi: 10.1186/1297-9686-17-2-265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renand G., and Krauss D.. 2002. Genetic relationship between fattening and slaughter traits in pure bred Charolais young bull. In: Proceedings of the 7th World Congress on Genetic Applied to Livestock Production; August 19–23, 2002 Montpellier, France. Communication no. 10-08. [Google Scholar]
- Richardson E. C., and Herd R. M.. 2004. Biological basis for variation in residual feed intake in beef cattle. 2. Synthesis of results following divergent selection. Aust. J. Exp. Agric. 44:431–440. doi: 10.1071/ea02221 [DOI] [Google Scholar]
- Richardson E. C., Herd R. M., Oddy V. H., Thompson J. M., Archer J. A., and Arthur P. F.. 2001. Body composition and implications for heat production of Angus steer progeny of parents selected for and against residual feed intake. Aust. J. Exp. Agric. 41:1065–1072. doi: 10.1071/ea00095 [DOI] [Google Scholar]
- Ríos Utrera A., and Van Vleck L. D.. 2004. Heritability estimates for carcass traits of cattle: a review. Genet. Mol. Res. 3:380–394. [PubMed] [Google Scholar]
- Robelin J., and Geay Y.. 1975. Estimation de la composition de la carcasse des taurillons à partir de la composition de la 6ème côte. Bull. Tech. CRZV Theix. 22:41–44. [Google Scholar]
- Robinson D. L., and Oddy V. H.. 2004. Genetic parameters for feed efficiency, fatness, muscle area and feeding behaviour of feedlot finished beef cattle. Livest. Prod. Sci. 90:255–270. doi: 10.1016/j.livprodsci.2004.06.011 [DOI] [Google Scholar]
- Rolf M. M., Taylor J. F., Schnabel R. D., McKay S. D., McClure M. C., Northcutt S. L., Kerley M. S., and Weaber R. L.. 2012. Genome-wide association analysis for feed efficiency in angus cattle. Anim. Genet. 43:367–374. doi: 10.1111/j.1365-2052.2011.02273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Beever J. E., Decker J. E., Faulkner D. B., Freetly H. C., Hansen S. L., Yampara-Iquise H., Johnson K. A., Kachman S. D., Kerley M. S., et al. . 2014. QTLs associated with dry matter intake, metabolic mid-test weight, growth and feed efficiency have little overlap across 4 beef cattle studies. BMC Genomics. 15:1004. doi: 10.1186/1471-2164-15-1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargolzaei M., Chesnais J. P., and Schenkel F. S.. 2014. A new approach for efficient genotype imputation using information from relatives. BMC Genomics. 15:478. doi: 10.1186/1471-2164-15-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabury C. M., Oldeschulte D. L., Saatchi M., Beever J. E., Decker J. E., Halley Y. A., Bhattarai E. K., Molaei M., Freetly H. C., Hansen S. L., et al. . 2017. Genome-wide association study for feed efficiency and growth traits in U.S. Beef cattle. BMC Genomics. 18:386. doi: 10.1186/s12864-017-3754-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer K. S., Turk P., Wagner W. R., and Felton E. E.. 2011. Residual feed intake, body composition, and fertility in yearling beef heifers. J. Anim. Sci. 89:1028–1034. doi: 10.2527/jas.2010-3322 [DOI] [PubMed] [Google Scholar]
- Sherman E. L., Nkrumah J. D., and Moore S. S.. 2010. Whole genome single nucleotide polymorphism associations with feed intake and feed efficiency in beef cattle. J. Anim. Sci. 88:16–22. doi: 10.2527/jas.2008-1759 [DOI] [PubMed] [Google Scholar]
- Shirali M., Varley P. F., and Jensen J.. 2018. Bayesian estimation of direct and correlated responses to selection on linear or ratio expressions of feed efficiency in pigs. Genet. Sel. Evol. 50:33. doi: 10.1186/s12711-018-0403-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. E., and Baldwin R. L.. 1974. Effects of breed, pregnancy, and lactation on weight of organs and tissues in dairy cattle. J. Dairy Sci. 57:1055–1060. doi: 10.3168/jds.S0022-0302(74)85008-3 [DOI] [Google Scholar]
- Sorbolini S., Bongiorni S., Cellesi M., Gaspa G., Dimauro C., Valentini A., and Macciotta N. P.. 2017. Genome wide association study on beef production traits in marchigiana cattle breed. J. Anim. Breed. Genet. 134:43–48. doi: 10.1111/jbg.12227 [DOI] [PubMed] [Google Scholar]
- Stucchi R., Plucińska G., Hummel J. J. A., Zahavi E. E., Guerra San Juan I., Klykov O., Scheltema R. A., Altelaar A. F. M., and Hoogenraad C. C.. 2018. Regulation of KIF1A-driven dense core vesicle transport: ca2+/cam controls DCV binding and liprin-α/TANC2 recruits dcvs to postsynaptic sites. Cell Rep. 24:685–700. doi: 10.1016/j.celrep.2018.06.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetens J., Thaller G., and Krattenmacher N.. 2014. Genetic and genomic dissection of dry matter intake at different lactation stages in primiparous holstein cows. J. Dairy Sci. 97:520–531. doi: 10.3168/jds.2013-7301 [DOI] [PubMed] [Google Scholar]
- Wozniak M. J., Melzer M., Dorner C., Haring H. U., and Lammers R.. 2005. The novel protein KBP regulates mitochondria localization by interaction with a kinesin-like protein. BMC Cell Biol. 6:35. doi: 10.1186/1471-2121-6-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., and Visscher P. M.. 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88:76–82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Hernandez-Sanabria E., and Guan L. L.. 2010. Characterization of variation in rumen methanogenic communities under different dietary and host feed efficiency conditions, as determined by PCR-denaturing gradient gel electrophoresis analysis. Appl. Environ. Microbiol. 76:3776–3786. doi: 10.1128/AEM.00010-10 [DOI] [PMC free article] [PubMed] [Google Scholar]